

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148404.8 - phase: 0
(416 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
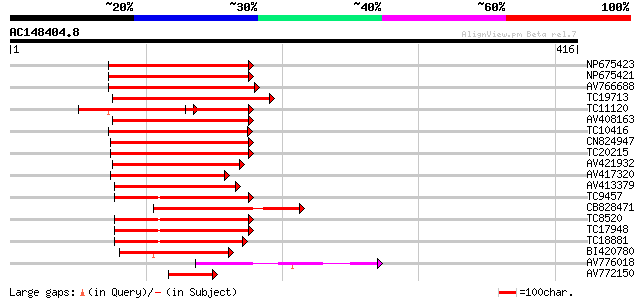
Score E
Sequences producing significant alignments: (bits) Value
NP675423 transcription factor MYB101 [Lotus japonicus] 154 3e-38
NP675421 transcription factor MYB103 [Lotus japonicus] 150 4e-37
AV766688 143 5e-35
TC19713 homologue to UP|P93474 (P93474) Myb26, partial (55%) 143 6e-35
TC11120 UP|Q84PP4 (Q84PP4) Transcription factor MYB102, complete 98 2e-34
AV408163 139 9e-34
TC10416 weakly similar to UP|O04108 (O04108) MYB factor, partial... 139 1e-33
CN824947 138 2e-33
TC20215 similar to AAS10104 (AAS10104) MYB transcription factor,... 135 2e-32
AV421932 127 3e-30
AV417320 126 6e-30
AV413379 121 2e-28
TC9457 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucros... 110 3e-25
CB828471 110 4e-25
TC8520 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucros... 107 3e-24
TC17948 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose... 107 4e-24
TC18881 similar to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucrose... 103 5e-23
BI420780 86 1e-17
AV776018 60 5e-10
AV772150 51 3e-07
>NP675423 transcription factor MYB101 [Lotus japonicus]
Length = 1047
Score = 154 bits (389), Expect = 3e-38
Identities = 68/107 (63%), Positives = 83/107 (77%)
Frame = +1
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
E GLKKGPWT ED ILV ++QK+G G+W ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 25 EIGLKKGPWTPEEDAILVDYIQKHGHGSWRALPKLAGLNRCGKSCRLRWTNYLRPDIKRG 204
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
T EEE+ II LH +GNKW+ +A LPGRTDNEIKNFWNT KK+
Sbjct: 205 KFTEEEEQLIINLHAVLGNKWSAIAGHLPGRTDNEIKNFWNTHLKKK 345
>NP675421 transcription factor MYB103 [Lotus japonicus]
Length = 924
Score = 150 bits (379), Expect = 4e-37
Identities = 63/107 (58%), Positives = 84/107 (77%)
Frame = +1
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
E G+KKGPW+ ED++LV ++QK+G G+W ++ + GL RCGKSCRLRW N+LRPD+++G
Sbjct: 25 ESGMKKGPWSQEEDKVLVDYIQKHGHGSWRALPQLAGLNRCGKSCRLRWTNYLRPDIKRG 204
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
+ EEE+ II LH +GNKWA +A+ LPGRTDNEIKN WNT KK+
Sbjct: 205 KFSDEEEQLIINLHASLGNKWATIASHLPGRTDNEIKNLWNTHLKKK 345
>AV766688
Length = 608
Score = 143 bits (361), Expect = 5e-35
Identities = 63/111 (56%), Positives = 82/111 (73%)
Frame = +2
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
+ GL+KG WTA ED L+A+V +YG NW + K GLARCGKSCRLRW N+LRPD+++G
Sbjct: 101 KSGLRKGTWTAEEDRKLIAYVTRYGCWNWRQLPKFAGLARCGKSCRLRWLNYLRPDIKRG 280
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRAN 183
T EEE II +H +GN+W+ +AA LPGRTDNE+KN W+T K+R + N
Sbjct: 281 HFTQEEEEIIIRMHKNLGNRWSIIAAELPGRTDNEVKNHWHTSLKRRVQQN 433
>TC19713 homologue to UP|P93474 (P93474) Myb26, partial (55%)
Length = 532
Score = 143 bits (360), Expect = 6e-35
Identities = 65/119 (54%), Positives = 86/119 (71%)
Frame = +3
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
++KGPWT ED IL+ ++ +GEG WN++ K GL R GKSCRLRW N+LRPD+R+G IT
Sbjct: 105 VRKGPWTIEEDLILINYIANHGEGVWNTLAKSAGLKRTGKSCRLRWLNYLRPDVRRGNIT 284
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRANLPIYPEDLSSE 194
EE+ I+ELH K GN+W+++A LPGRTDNEIKNFW TR +K + + + SSE
Sbjct: 285 PEEQLLIMELHAKWGNRWSKIAKHLPGRTDNEIKNFWRTRIQKHIKQAESLQQQQSSSE 461
>TC11120 UP|Q84PP4 (Q84PP4) Transcription factor MYB102, complete
Length = 1281
Score = 98.2 bits (243), Expect(2) = 2e-34
Identities = 47/92 (51%), Positives = 64/92 (69%), Gaps = 4/92 (4%)
Frame = +1
Query: 51 RQSILSA-DQETDEGDTTGGEG---GEGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRK 106
R++I++A DQ ++ T G E GLKKGPWT ED+ LV H+QK+G G+W ++ K
Sbjct: 13 RENIITALDQVSN*VTHTMGRSPCCDENGLKKGPWTPEEDQKLVEHIQKHGHGSWRALPK 192
Query: 107 CTGLARCGKSCRLRWANHLRPDLRKGAITAEE 138
GL RCGKSCRLRW N+LRPD+++G + EE
Sbjct: 193 LAGLNRCGKSCRLRWTNYLRPDIKRGKFSPEE 288
Score = 64.7 bits (156), Expect(2) = 2e-34
Identities = 28/50 (56%), Positives = 36/50 (72%)
Frame = +2
Query: 130 RKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
R G ++E+ I+ LH +GNKW+ +A LPGRTDNEIKNFWNT KK+
Sbjct: 263 RGGNFLQKKEQTILHLHSILGNKWSTIATHLPGRTDNEIKNFWNTHLKKK 412
>AV408163
Length = 425
Score = 139 bits (350), Expect = 9e-34
Identities = 58/104 (55%), Positives = 79/104 (75%)
Frame = +2
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
+ KG WT ED+ L+++++ +GEG W S+ K GL RCGKSCRLRW N+LRPDL++G T
Sbjct: 110 INKGAWTKEEDDRLISYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRPDLKRGNFT 289
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
EE+ II+LH +GNKW+ +A LPGRTDNEIKN+WNT +++
Sbjct: 290 EEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRK 421
>TC10416 weakly similar to UP|O04108 (O04108) MYB factor, partial (41%)
Length = 1046
Score = 139 bits (349), Expect = 1e-33
Identities = 59/106 (55%), Positives = 81/106 (75%)
Frame = +2
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
+ G++KG WTA ED+ L+A+V +YG NW + K GLARCGKSCRLRW N+LRP++++G
Sbjct: 116 KSGMRKGTWTAEEDKKLIAYVTRYGCWNWRQLPKFAGLARCGKSCRLRWLNYLRPNIKRG 295
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
T EEE II +H K+GN+W+ +AA LPGRTD+E+KN W+T K+
Sbjct: 296 NFTQEEEEIIIRMHKKLGNRWSTIAAELPGRTDSEVKNHWHTTLKR 433
>CN824947
Length = 738
Score = 138 bits (347), Expect = 2e-33
Identities = 59/105 (56%), Positives = 80/105 (76%)
Frame = +1
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLK+G WTA EDE+L ++Q GEG+W S+ K GL RCGKSCRLRW N+LR DL++G I
Sbjct: 181 GLKRGRWTAEEDELLTKYIQASGEGSWRSLPKNAGLLRCGKSCRLRWINYLRGDLKRGNI 360
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
+ EEE I++LH GN+W+ +A+ +PGRTDNEIKN+WN+ ++
Sbjct: 361 SDEEESLIVKLHASFGNRWSLIASHMPGRTDNEIKNYWNSHLSRK 495
>TC20215 similar to AAS10104 (AAS10104) MYB transcription factor, partial
(35%)
Length = 464
Score = 135 bits (339), Expect = 2e-32
Identities = 57/105 (54%), Positives = 81/105 (76%)
Frame = +2
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLK+G W+ ED+IL +++ GEG+W S+ K GL RCGKSCRLRW N+LR D+++G I
Sbjct: 59 GLKRGRWSEEEDKILTDFIKQNGEGSWKSLPKNAGLLRCGKSCRLRWINYLREDVKRGNI 238
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
T+EEE I++LH +GN+W+ +A LPGRTDNEIKN+WN+ +++
Sbjct: 239 TSEEEEIIVKLHAALGNRWSVIAGHLPGRTDNEIKNYWNSHLRRK 373
>AV421932
Length = 402
Score = 127 bits (320), Expect = 3e-30
Identities = 55/97 (56%), Positives = 73/97 (74%)
Frame = +3
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
+K+G W+ EDE L+ ++ +G G W+ V + GL RCGKSCRLRW N+LRPD+R+G T
Sbjct: 111 VKRGLWSPEEDEKLIRYITTHGYGCWSEVPEKAGLQRCGKSCRLRWINYLRPDIRRGRFT 290
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFW 172
EEE+ II LH +GN+WA +A+ LPGRTDNEIKN+W
Sbjct: 291 PEEEKLIISLHGVVGNRWAHIASHLPGRTDNEIKNYW 401
>AV417320
Length = 369
Score = 126 bits (317), Expect = 6e-30
Identities = 52/87 (59%), Positives = 71/87 (80%)
Frame = +3
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLKKGPWT+ ED+IL++++QK+G GNW ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 108 GLKKGPWTSEEDQILISYIQKHGHGNWRALPKHAGLLRCGKSCRLRWINYLRPDIKRGNF 287
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLP 161
TAEEE II++H +GN+W+ +AA LP
Sbjct: 288 TAEEEELIIKMHELLGNRWSAIAAKLP 368
>AV413379
Length = 416
Score = 121 bits (303), Expect = 2e-28
Identities = 53/92 (57%), Positives = 69/92 (74%)
Frame = +3
Query: 78 KGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITAE 137
KG WT ED+ L+A+++ +GEG W S+ K GL RCGKSCRLRW N+LRPDL++G T +
Sbjct: 141 KGAWTKEEDDRLIAYIRAHGEGCWRSLPKSAGLLRCGKSCRLRWINYLRPDLKRGNFTPQ 320
Query: 138 EERRIIELHHKMGNKWAQMAALLPGRTDNEIK 169
E+ II+LH +GNKW+ +A L GRTDNEIK
Sbjct: 321 EDELIIKLHSLLGNKWSLIAGRLAGRTDNEIK 416
>TC9457 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and
sucrose-responsive element binding factor, partial (43%)
Length = 860
Score = 110 bits (276), Expect = 3e-25
Identities = 53/102 (51%), Positives = 65/102 (62%)
Frame = +3
Query: 78 KGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITAE 137
KGPW+ EDE L V+K+G NW+ + K R GKSCRLRW N L P + A TAE
Sbjct: 78 KGPWSPEEDEALQRLVEKHGPRNWSLISKSIP-GRSGKSCRLRWCNQLSPQVEHRAFTAE 254
Query: 138 EERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
E+ II H + GNKWA +A LL GRTDN IKN WN+ K++
Sbjct: 255 EDDTIIRAHARFGNKWATIARLLSGRTDNAIKNHWNSTLKRK 380
>CB828471
Length = 525
Score = 110 bits (275), Expect = 4e-25
Identities = 53/111 (47%), Positives = 71/111 (63%)
Frame = +1
Query: 106 KCTGLARCGKSCRLRWANHLRPDLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTD 165
K GL RCGKSCRLRWAN+LRPD+++G + EEE II LH +GN+W+ +AA LPGRTD
Sbjct: 4 KQAGLLRCGKSCRLRWANYLRPDIKRGNFSREEEDEIINLHELLGNRWSAIAARLPGRTD 183
Query: 166 NEIKNFWNTRCKKRGRANLPIYPEDLSSECLLNQENADMLTNEASQHDEAE 216
NEIKN W+T KKR + P+ + L+ + + + +H E E
Sbjct: 184 NEIKNVWHTHLKKR------LQPQTKQKQTNLDLDASKSNKDAKKEHQEDE 318
>TC8520 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and
sucrose-responsive element binding factor, partial (36%)
Length = 1778
Score = 107 bits (268), Expect = 3e-24
Identities = 51/102 (50%), Positives = 64/102 (62%)
Frame = +1
Query: 78 KGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITAE 137
KGPW+ EDE L V+++G NW+ + K R GKSCRLRW N L P + A T E
Sbjct: 184 KGPWSPEEDEALQKLVERHGPRNWSLISKSIP-GRSGKSCRLRWCNQLSPQVEHRAFTPE 360
Query: 138 EERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
E+ II H + GNKWA +A LL GRTDN IKN WN+ K++
Sbjct: 361 EDDTIIRAHARFGNKWATIARLLSGRTDNAIKNHWNSTLKRK 486
>TC17948 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose-responsive
element binding factor, partial (50%)
Length = 487
Score = 107 bits (267), Expect = 4e-24
Identities = 48/102 (47%), Positives = 65/102 (63%)
Frame = +2
Query: 78 KGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITAE 137
KGPW+A ED IL V+++G NW+ + + R GKSCRLRW N L P + + +
Sbjct: 170 KGPWSAEEDRILTRLVERHGARNWSLISRYIK-GRSGKSCRLRWCNQLSPTVEHRPFSVQ 346
Query: 138 EERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
E+ II H + GN+WA +A LLPGRTDN +KN WN+ K+R
Sbjct: 347 EDETIIAAHARFGNRWAAIARLLPGRTDNAVKNHWNSTLKRR 472
>TC18881 similar to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucrose-responsive
element binding factor, partial (28%)
Length = 346
Score = 103 bits (257), Expect = 5e-23
Identities = 48/97 (49%), Positives = 60/97 (61%)
Frame = +1
Query: 78 KGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITAE 137
KGPW+ EDE L VQ +G NW ++ K R GKSCRLRW N L P++ T E
Sbjct: 55 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIP-GRSGKSCRLRWCNQLSPEVEHRPFTPE 231
Query: 138 EERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNT 174
E+ II H + GNKWA +A +L GRTDN +KN WN+
Sbjct: 232 EDSTIIRAHARCGNKWATIARILNGRTDNAVKNHWNS 342
>BI420780
Length = 475
Score = 85.9 bits (211), Expect = 1e-17
Identities = 42/86 (48%), Positives = 58/86 (66%), Gaps = 2/86 (2%)
Frame = +1
Query: 81 WTAAEDEILVAHVQKYGEGNWNSV--RKCTGLARCGKSCRLRWANHLRPDLRKGAITAEE 138
W++ ED +L A+VQ+YG WN V R T L R KSC RW N+L+P ++KG++T EE
Sbjct: 211 WSSEEDALLHAYVQQYGPREWNLVSQRMNTPLNRDTKSCLERWKNYLKPGIKKGSLTKEE 390
Query: 139 ERRIIELHHKMGNKWAQMAALLPGRT 164
+R +I L GNKW ++AA +PGRT
Sbjct: 391 QRLVILLQANYGNKWKKIAAEVPGRT 468
>AV776018
Length = 476
Score = 60.5 bits (145), Expect = 5e-10
Identities = 43/141 (30%), Positives = 60/141 (42%), Gaps = 4/141 (2%)
Frame = +1
Query: 137 EEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRANLPIYPEDLSSECL 196
EE+ IIELH +GN+W+Q A PGRTDNEIKN WN+ KK+
Sbjct: 22 EEDNLIIELHAVLGNRWSQDATQWPGRTDNEIKNLWNSCLKKK----------------- 150
Query: 197 LNQENADMLT----NEASQHDEAENKVPEVIFKDYKLRPDILPPCFDKLLAGLLLRPSKR 252
L Q D +T +E E +NK E + + L
Sbjct: 151 LRQRGIDPVTHKPLSEVENGKEDKNKSQEKVSNELNLL-------------------KSE 273
Query: 253 PWKSDMNLYSYSNNAAAPAAF 273
+KSD Y ++++ AP A+
Sbjct: 274 SFKSDAASYEQNSSSLAPKAY 336
>AV772150
Length = 535
Score = 51.2 bits (121), Expect = 3e-07
Identities = 21/36 (58%), Positives = 28/36 (77%)
Frame = +3
Query: 117 CRLRWANHLRPDLRKGAITAEEERRIIELHHKMGNK 152
CRLRW N+LRPDL+KG + +EE II+LH +GN+
Sbjct: 27 CRLRWINYLRPDLKKGQLAEDEEDLIIKLHALLGNR 134
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.131 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,308,300
Number of Sequences: 28460
Number of extensions: 130458
Number of successful extensions: 784
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 767
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 776
length of query: 416
length of database: 4,897,600
effective HSP length: 93
effective length of query: 323
effective length of database: 2,250,820
effective search space: 727014860
effective search space used: 727014860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148404.8


