

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148396.11 + phase: 0
(745 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
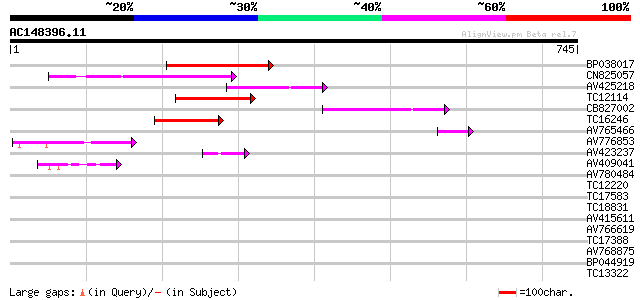
Score E
Sequences producing significant alignments: (bits) Value
BP038017 134 4e-32
CN825057 123 9e-29
AV425218 110 8e-25
TC12114 weakly similar to GB|BAC16510.1|23237937|AP005198 transp... 97 9e-21
CB827002 89 3e-18
TC16246 weakly similar to UP|Q8S2M2 (Q8S2M2) Far-red impaired re... 79 2e-15
AV765466 48 5e-06
AV776853 45 4e-05
AV423237 43 2e-04
AV409041 41 8e-04
AV780484 37 0.009
TC12220 37 0.015
TC17583 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired re... 32 0.37
TC18831 similar to UP|Q9LI87 (Q9LI87) Emb|CAB12631.1, partial (44%) 31 0.82
AV415611 30 1.1
AV766619 28 4.1
TC17388 28 5.3
AV768875 27 9.1
BP044919 27 9.1
TC13322 similar to UP|Q9ZS13 (Q9ZS13) RNA helicase (Fragment), p... 27 9.1
>BP038017
Length = 570
Score = 134 bits (338), Expect = 4e-32
Identities = 63/141 (44%), Positives = 89/141 (62%)
Frame = +2
Query: 206 YSRNLKELIKDSDADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLF 265
Y ++ ++ DA ++ F++ + NP F+Y + D+E ++ +VFWAD R Y+ F
Sbjct: 131 YVKSSQKRTLGRDAQNLLNYFKKMQGKNPGFYYAIQLDDENRMINVFWADARSRSAYNYF 310
Query: 266 GDVVSFDTTYRTNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGG 325
GD V FDT YR N+Y + FAPFTG+NHH Q++ FG ALL +E E SF WLF T+L AM
Sbjct: 311 GDAVIFDTMYRPNQYQVPFAPFTGVNHHGQNVLFGCALLLDESESSFTWLFRTWLSAMND 490
Query: 326 HKPVMIITDQDGGMKNAIGAV 346
PV I TDQD ++ A+ V
Sbjct: 491 RPPVSITTDQDRAIQAAVAQV 553
>CN825057
Length = 721
Score = 123 bits (309), Expect = 9e-29
Identities = 80/249 (32%), Positives = 124/249 (49%), Gaps = 3/249 (1%)
Frame = +1
Query: 52 FYKAYAYVAGF--SVRNSIKTKDKDGVKWKYFLCSKEGFKEEKKVDKPQLLIAENSLSKS 109
FY+ YA GF S++NS ++K F CS+ G + E+ +
Sbjct: 10 FYQEYAKSMGFTTSIKNSRRSKKTKEFIDAKFACSRYG------------VTPESDGGSN 153
Query: 110 RKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLVSPSKRQFLRSARNVTSVHKNIL 169
R+ + + CKA + +KR DGK+ + F + H+H L+ P+ R RNV KN +
Sbjct: 154 RRSSVKKTDCKACMHVKRKPDGKWIIHEFIKEHNHELL-PALAYHFRIHRNVKLAEKNNM 330
Query: 170 FSCNRANVGTSKSYQIMKEQVGSYENIGCTQRDLQNYSRNLKELIKDS-DADMFIDNFRR 228
+ + T K Y M Q G NI DL + + + L D DA + ++ F+
Sbjct: 331 DILHAVSERTRKMYVEMSRQSGGCLNIESLVGDLNDQFKKGQYLAMDEGDAQVMLEYFKH 510
Query: 229 KREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKYFMIFAPFT 288
++ NP+FFY + + E +L+++FW D +Y F DVVSFDT+Y + + FAPF
Sbjct: 511 IQKENPNFFYSIDLNEEQRLRNIFWIDAKSINDYLSFNDVVSFDTSYIKSNEKLPFAPFV 690
Query: 289 GINHHRQSI 297
G+NHH Q I
Sbjct: 691 GVNHHCQPI 717
>AV425218
Length = 419
Score = 110 bits (275), Expect = 8e-25
Identities = 53/133 (39%), Positives = 78/133 (57%)
Frame = +2
Query: 285 APFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMKNAIG 344
A G+NHH QS+ FG ALL +E ESFVWLF++ L M G P IITD MK AI
Sbjct: 11 ATLVGVNHHGQSVLFGCALLSSEDSESFVWLFQSLLHCMSGVPPQGIITDHSEAMKKAIE 190
Query: 345 AVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSCVWNSESSEEFDLEWNNIISD 404
VL + HR+C+ +I+KKL +K+ + ++ V+++ +EF+ W I+ D
Sbjct: 191 TVLPSTRHRWCLSYIMKKLPQKL-LGYAQYESIRHHLQNVVYDAVVIDEFERNWKKIVED 367
Query: 405 YSLEGNGWLSTMY 417
+ LE N WL+ ++
Sbjct: 368 FGLEDNEWLNELF 406
>TC12114 weakly similar to GB|BAC16510.1|23237937|AP005198 transposase-like
{Oryza sativa (japonica cultivar-group);}, partial (8%)
Length = 488
Score = 97.1 bits (240), Expect = 9e-21
Identities = 46/105 (43%), Positives = 68/105 (63%)
Frame = +1
Query: 218 DADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRT 277
+A + F+RK N F++ Y+ D+E ++ +VFW D +Y FGD+VS D+TY T
Sbjct: 43 EAGYILQYFQRKLVENSPFYHAYQLDDEDQITNVFWVDARMLIDYGYFGDMVSLDSTYCT 222
Query: 278 NKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKA 322
+ A F+G NHHR+++ FGAALL +E ES+ WLFE+FL+A
Sbjct: 223 HSSNRPLAVFSGFNHHRKAVIFGAALLYDETTESY*WLFESFLEA 357
>CB827002
Length = 534
Score = 89.0 bits (219), Expect = 3e-18
Identities = 56/168 (33%), Positives = 90/168 (53%), Gaps = 1/168 (0%)
Frame = +1
Query: 412 WLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESENSFFGNYLNHNLTLVEFWVRFDSAL 471
WLS +Y P + +DTF A + T RS+S NS+F Y+N + L +F+ ++ AL
Sbjct: 40 WLS-LYSSCRQGAPVHLRDTFFAE-MSITQRSDSMNSYFDGYVNASTNLNQFFKLYEKAL 213
Query: 472 AAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKELWSACVDCGIEGTK 531
++ KE+ AD +T+++ P L +EK A E+YT + F FQ+EL +
Sbjct: 214 ESRNEKEVRADYDTMNTLPVLRTPSPMEKQASELYTRKIFMRFQEELVGTLTFMASK-AD 390
Query: 532 EEGENLSFSILD-NAVRKHREVVYCLSNNIAHCSCKMFESEGIPCRHI 578
++GE +++ + K V + + A CSC+MFE G+ CRHI
Sbjct: 391 DDGEVITYHVAKFGEDHKAYYVKFNVLEMKASCSCQMFEFSGLLCRHI 534
>TC16246 weakly similar to UP|Q8S2M2 (Q8S2M2) Far-red impaired response
protein-like, partial (4%)
Length = 660
Score = 79.3 bits (194), Expect = 2e-15
Identities = 34/91 (37%), Positives = 60/91 (65%), Gaps = 1/91 (1%)
Frame = +2
Query: 191 GSYENIGCTQRDLQNY-SRNLKELIKDSDADMFIDNFRRKREINPSFFYDYEADNEGKLK 249
G +E++G T+ DL N+ ++ +E I++ DA + K + +P FFY + + L+
Sbjct: 65 GGHESLGFTKTDLSNHIAKPKRERIQNGDAAAALSYLEGKADNDPMFFYKFTKTGDESLE 244
Query: 250 HVFWADGICRKNYSLFGDVVSFDTTYRTNKY 280
++FW DG+ R +Y++FGDV++FD+TY+ NKY
Sbjct: 245 NLFWCDGVSRMDYNVFGDVIAFDSTYKKNKY 337
>AV765466
Length = 599
Score = 48.1 bits (113), Expect = 5e-06
Identities = 19/47 (40%), Positives = 28/47 (59%)
Frame = -2
Query: 563 CSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTKLATSKPIF 609
C C++FE GI CRH L IL + E+P Y+++RW K K ++
Sbjct: 592 CDCRLFEFRGILCRHSLAILSQERVKEVPDRYVLDRWRKNMRRKYVY 452
>AV776853
Length = 625
Score = 45.1 bits (105), Expect = 4e-05
Identities = 45/181 (24%), Positives = 75/181 (40%), Gaps = 18/181 (9%)
Frame = +1
Query: 4 DDNEVDFS-------SPSKDGND-PKNTSVEWIPACEDELKPVIGKVFDTL--------V 47
D+ +V+F S S + ND E + A +D K ++ D +
Sbjct: 79 DEGDVEFEYDISSGDSGSGEHNDLGDKDEAEDVDAKDDHYKRILDLSADDIRHLDFGSEE 258
Query: 48 EGGDFYKAYAYVAGFSVRNSIKTKDKDG-VKWKYFLCSKEGFKEEKKVDKPQLLIAENSL 106
E FY+AYA GF VR +D G V + F+C+++G + +K ++
Sbjct: 259 EAYQFYQAYAKYQGFIVRKDDIGRDYHGNVNMRQFVCNRQGLRSKKHYNRTD-------- 414
Query: 107 SKSRKRKLTREGCKARLVLKRTID-GKYEVSNFYEGHSHGLVSPSKRQFLRSARNVTSVH 165
K + +T C A+L + GK++V +F E H+H L F+ R +
Sbjct: 415 RKRDHKPVTHTNCLAKLRVHLDYKIGKWKVVSFEECHNHELTPARFVHFIPPYRVMNDAD 594
Query: 166 K 166
K
Sbjct: 595 K 597
>AV423237
Length = 310
Score = 43.1 bits (100), Expect = 2e-04
Identities = 25/62 (40%), Positives = 31/62 (49%)
Frame = +2
Query: 254 ADGICRKNYSLFGDVVSFDTTYRTNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFV 313
AD R NYS FGD V FDTT + + G + + + AL+ NE E SFV
Sbjct: 14 ADATSRMNYSYFGDAVIFDTT--IDNHIESICSSWG*SSWATCVIWLVALIANESESSFV 187
Query: 314 WL 315
WL
Sbjct: 188 WL 193
>AV409041
Length = 349
Score = 40.8 bits (94), Expect = 8e-04
Identities = 36/120 (30%), Positives = 58/120 (48%), Gaps = 9/120 (7%)
Frame = +3
Query: 37 PVIGKVFDTLVEGGD----FYKAYAYVAGF-----SVRNSIKTKDKDGVKWKYFLCSKEG 87
PV+ +D E + FYK YA AGF S R S +K+ K F C + G
Sbjct: 21 PVLEPHYDIEFESHEAAYAFYKEYAKSAGFGTAKLSSRRSRASKEFIDAK---FSCIRYG 191
Query: 88 FKEEKKVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLV 147
K++ ++++++ K+ GCKA + +KR DGK+ V +F + H+H L+
Sbjct: 192 NKQQ----------SDDAINPRPSPKI---GCKASMHVKRRQDGKWYVYSFVKEHNHELL 332
>AV780484
Length = 529
Score = 37.4 bits (85), Expect = 0.009
Identities = 21/54 (38%), Positives = 26/54 (47%)
Frame = -1
Query: 233 NPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKYFMIFAP 286
NP+FFY + D L VFW D R +Y +V T Y NKY + F P
Sbjct: 166 NPNFFYAIDLDLNRHLTSVFWVDIKGRLDYETSMMLVLIHTHYLKNKYKIPFFP 5
>TC12220
Length = 473
Score = 36.6 bits (83), Expect = 0.015
Identities = 16/35 (45%), Positives = 22/35 (62%)
Frame = +1
Query: 569 ESEGIPCRHILFILKGKGFSEIPSHYIVNRWTKLA 603
E EG+ CRH L + E+PS YI++RWT+ A
Sbjct: 1 EYEGVLCRHPLRVFXILELREVPSRYILHRWTRNA 105
>TC17583 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired response
protein, mutator-like transposase-like protein,
phytochrome A signaling protein-like, partial (3%)
Length = 666
Score = 32.0 bits (71), Expect = 0.37
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +3
Query: 261 NYSLFGDVVSFDTTYRTNKYF 281
+Y FGDVVS DTTY TN +
Sbjct: 15 DYEYFGDVVSLDTTYSTNNAY 77
>TC18831 similar to UP|Q9LI87 (Q9LI87) Emb|CAB12631.1, partial (44%)
Length = 700
Score = 30.8 bits (68), Expect = 0.82
Identities = 18/80 (22%), Positives = 34/80 (42%)
Frame = +3
Query: 385 VWNSESSEEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSE 444
VWNS S + ++ N ++++ + G S ++ + S+W ++
Sbjct: 138 VWNSRSEQSPGVDKMNALANFLWKNGGPRSRLHLIHSVW---------------ANFQTS 272
Query: 445 SENSFFGNYLNHNLTLVEFW 464
S N FGN H + +FW
Sbjct: 273 SNNIIFGNRWRHLIGERDFW 332
>AV415611
Length = 219
Score = 30.4 bits (67), Expect = 1.1
Identities = 10/24 (41%), Positives = 16/24 (66%)
Frame = +3
Query: 563 CSCKMFESEGIPCRHILFILKGKG 586
CSCK ++ G+PC H + ++ G G
Sbjct: 132 CSCKTWQLTGVPCCHAIAVIVGIG 203
>AV766619
Length = 412
Score = 28.5 bits (62), Expect = 4.1
Identities = 19/105 (18%), Positives = 45/105 (42%), Gaps = 3/105 (2%)
Frame = +2
Query: 327 KPVMIITDQDGGMKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSG---FNDRFKS 383
+P+ + + G+K ++ + + H +C+ ++ +KL++ + + ND + +
Sbjct: 14 EPITFVANCQNGLKKSLSEIFEKCHHSYCLRYLAEKLNKDLKGQFSHEARRFMINDFYAA 193
Query: 384 CVWNSESSEEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYF 428
+ E F+ N + S E W+ M + W A+F
Sbjct: 194 AY--APKQETFERSVEN-MKGISSEAFNWV--MQNEPEHWANAFF 313
>TC17388
Length = 500
Score = 28.1 bits (61), Expect = 5.3
Identities = 14/46 (30%), Positives = 24/46 (51%)
Frame = +1
Query: 536 NLSFSILDNAVRKHREVVYCLSNNIAHCSCKMFESEGIPCRHILFI 581
N FS VRK C S+++ +C+C + + C+HI+F+
Sbjct: 91 NGGFSHH*TGVRKIATKAKCFSHSVFNCAC*L-----VTCKHIIFV 213
>AV768875
Length = 232
Score = 27.3 bits (59), Expect = 9.1
Identities = 12/50 (24%), Positives = 21/50 (42%)
Frame = -3
Query: 592 SHYIVNRWTKLATSKPIFDCDGNVLEACSKFESESTLVSKTWSQLLKCMH 641
S + RW +L + P C+GN + S S+ + + +C H
Sbjct: 158 SRTVAKRWARLGWNSPPLTCEGNKISFMSPIRSQGIEIEARMVE--RCCH 15
>BP044919
Length = 519
Score = 27.3 bits (59), Expect = 9.1
Identities = 13/54 (24%), Positives = 32/54 (59%)
Frame = +3
Query: 642 MAGTNKEKLLLIYDEGCSIELKMSKMKTDVVSRPLDELEAFIGSNVAKTIEVLP 695
+AGT+ + ++++ +G +L ++ + V+S+ + EAF+ S+V + +P
Sbjct: 87 IAGTDDKPMIVVKHKGEEKKLVPEEISSMVLSKMKEIAEAFLESSVKNAVVTVP 248
>TC13322 similar to UP|Q9ZS13 (Q9ZS13) RNA helicase (Fragment), partial (7%)
Length = 528
Score = 27.3 bits (59), Expect = 9.1
Identities = 12/31 (38%), Positives = 16/31 (50%)
Frame = +3
Query: 614 NVLEACSKFESESTLVSKTWSQLLKCMHMAG 644
NV+E C FE S + S+ W LL + G
Sbjct: 432 NVMECCKGFEKPSPIQSRAWPFLLDGRDLIG 524
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.134 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,850,898
Number of Sequences: 28460
Number of extensions: 198036
Number of successful extensions: 954
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 946
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 948
length of query: 745
length of database: 4,897,600
effective HSP length: 97
effective length of query: 648
effective length of database: 2,136,980
effective search space: 1384763040
effective search space used: 1384763040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148396.11


