

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148360.5 + phase: 0
(315 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
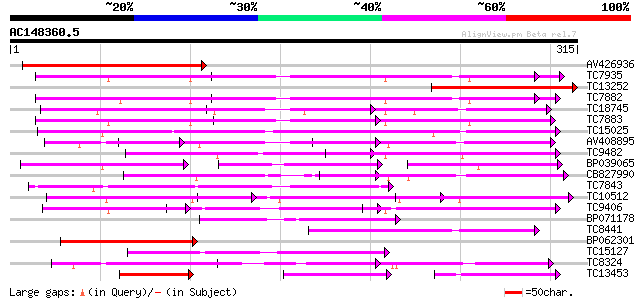
Score E
Sequences producing significant alignments: (bits) Value
AV426936 169 4e-43
TC7935 similar to UP|O49447 (O49447) ADP, ATP carrier-like prote... 135 7e-33
TC13252 similar to UP|Q9MA27 (Q9MA27) T5E21.6, partial (17%) 128 1e-30
TC7882 homologue to UP|O49875 (O49875) Adenine nucleotide transl... 124 3e-29
TC18745 weakly similar to UP|ADT2_HUMAN (P05141) ADP,ATP carrier... 114 3e-26
TC7883 homologue to UP|ADT1_GOSHI (O22342) ADP,ATP carrier prote... 108 2e-24
TC15025 similar to UP|MCAT_ARATH (Q93XM7) Mitochondrial carnitin... 89 1e-18
AV408895 89 1e-18
TC9482 similar to UP|Q9FI73 (Q9FI73) Mitochondrial carrier prote... 72 1e-13
BP039065 69 1e-12
CB827990 67 4e-12
TC7843 weakly similar to GB|AAF03412.1|6090963|AF188712 mitochon... 65 1e-11
TC10512 similar to UP|Q9FI73 (Q9FI73) Mitochondrial carrier prot... 64 4e-11
TC9406 similar to UP|Q9M058 (Q9M058) Ca-dependent solute carrier... 63 7e-11
BP071178 60 4e-10
TC8441 homologue to UP|ADT2_SOLTU (P27081) ADP,ATP carrier prote... 59 8e-10
BP062301 59 1e-09
TC15127 weakly similar to GB|AAG29586.1|11094343|AF155813 mitoch... 53 8e-08
TC8324 UP|Q8VWY1 (Q8VWY1) Mitochondrial phosphate transporter, c... 52 1e-07
TC13453 similar to UP|Q9ZNY4 (Q9ZNY4) Mitochondrial energy trans... 50 4e-07
>AV426936
Length = 401
Score = 169 bits (429), Expect = 4e-43
Identities = 81/102 (79%), Positives = 92/102 (89%)
Frame = +2
Query: 8 ILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEG 67
+LDH+P+FAKELLAGG+AGGFAK+VVAPLER+KIL QTRR EF+S+GL GS RIAKTEG
Sbjct: 95 LLDHVPVFAKELLAGGIAGGFAKSVVAPLERVKILLQTRRAEFQSSGLWGSATRIAKTEG 274
Query: 68 LLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWK 109
LGFYRGNGASVARIIPYA LH+MSYEEYRR I+ FP+VWK
Sbjct: 275 FLGFYRGNGASVARIIPYAALHYMSYEEYRRWIVLTFPDVWK 400
Score = 38.5 bits (88), Expect = 0.001
Identities = 26/94 (27%), Positives = 41/94 (42%)
Frame = +2
Query: 114 DLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK 173
+L+AG ++GG A PL+ ++ L + E G+ ++ K
Sbjct: 125 ELLAGGIAGGFAKSVVAPLERVKILLQTR-------------RAEFQSSGLWGSATRIAK 265
Query: 174 EGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKR 207
G G YRG ++ I PYA L + YEE +R
Sbjct: 266 TEGFLGFYRGNGASVARIIPYAALHYMSYEEYRR 367
Score = 35.4 bits (80), Expect = 0.013
Identities = 21/95 (22%), Positives = 44/95 (46%)
Frame = +2
Query: 212 DYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQK 271
D+ +L G +AG ++ PLE V+ +Q + E + G S IA+
Sbjct: 101 DHVPVFAKELLAGGIAGGFAKSVVAPLERVKILLQTRR----AEFQSSGLWGSATRIAKT 268
Query: 272 QGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
+G+ + G + +++P AA+ + Y+ + ++
Sbjct: 269 EGFLGFYRGNGASVARIIPYAALHYMSYEEYRRWI 373
>TC7935 similar to UP|O49447 (O49447) ADP, ATP carrier-like protein,
partial (83%)
Length = 1465
Score = 135 bits (341), Expect = 7e-33
Identities = 89/290 (30%), Positives = 140/290 (47%), Gaps = 10/290 (3%)
Frame = +3
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSA-------GLSGSVRRIAKTEG 67
F + L GG++ +KT AP+ER+K+L Q + +S G+ R K EG
Sbjct: 243 FLVDFLMGGVSAAVSKTAAAPIERIKLLIQNQDEMIKSGRLSEPYKGIGDCFARTTKDEG 422
Query: 68 LLGFYRGNGASVARIIPYAGLHFMSYEEYRRL--IMQAFPNVWKGPTLDLMAGSLSGGTA 125
+ +RGN A+V R P L+F + ++RL + WK +L +G +G ++
Sbjct: 423 AIALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAAGASS 602
Query: 126 VLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVA 185
+LF Y LD RT+LA S K E+ + G+ D KT + GI GLYRG +
Sbjct: 603 LLFVYSLDYARTRLANDSKSAKK-------GGERQFNGLVDVYKKTIQSDGIAGLYRGFS 761
Query: 186 PTLFGIFPYAGLKFYFYEEMKRRV-PEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQ 244
+ GI Y GL F Y+ +K V D + S A G + +YP++ VRR+
Sbjct: 762 ISCIGIIVYRGLYFGMYDSLKPVVLVGDMQDSFFASFLLGWGITIGAGLASYPIDTVRRR 941
Query: 245 MQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAI 294
M + + E + K ++ + +I K+G K+LF G N ++ V A +
Sbjct: 942 MM---MTSGEAVKYKSSLDAFKVIIAKEGTKSLFKGAGANILRAVAGAGV 1082
Score = 67.8 bits (164), Expect = 2e-12
Identities = 51/203 (25%), Positives = 90/203 (44%), Gaps = 7/203 (3%)
Frame = +3
Query: 113 LDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 172
+D + G +S + P++ I+ + Q ++ SG ++ + Y+GI DC ++T
Sbjct: 249 VDFLMGGVSAAVSKTAAAPIERIKLLIQNQ----DEMIKSGRLS--EPYKGIGDCFARTT 410
Query: 173 KEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKR-----RVPEDYKKSIMAKLTCGSVA 227
K+ G L+RG + FP L F F + KR + + Y K L G A
Sbjct: 411 KDEGAIALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAA 590
Query: 228 GLLGQTFTYPLEVVRRQMQVQNLAASE--EAELKGTMRSMVLIAQKQGWKTLFSGLSINY 285
G F Y L+ R ++ + +A + E + G + Q G L+ G SI+
Sbjct: 591 GASSLLFVYSLDYARTRLANDSKSAKKGGERQFNGLVDVYKKTIQSDGIAGLYRGFSISC 770
Query: 286 IKVVPSAAIGFTVYDTMKSYLRV 308
I ++ + F +YD++K + V
Sbjct: 771 IGIIVYRGLYFGMYDSLKPVVLV 839
>TC13252 similar to UP|Q9MA27 (Q9MA27) T5E21.6, partial (17%)
Length = 638
Score = 128 bits (322), Expect = 1e-30
Identities = 63/81 (77%), Positives = 70/81 (85%)
Frame = +2
Query: 235 TYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAI 294
TYPL VVRRQMQV L+AS+ ELKGT+RSMVLIA K GWK LFSGLSINY+KVVPS A+
Sbjct: 2 TYPLAVVRRQMQVPILSASDNVELKGTLRSMVLIAHKHGWKQLFSGLSINYLKVVPSVAL 181
Query: 295 GFTVYDTMKSYLRVPSRDEVD 315
GFTVYDTMK YLRVPSR+E +
Sbjct: 182 GFTVYDTMKLYLRVPSREEAE 244
>TC7882 homologue to UP|O49875 (O49875) Adenine nucleotide translocator,
complete
Length = 1656
Score = 124 bits (310), Expect = 3e-29
Identities = 84/290 (28%), Positives = 138/290 (46%), Gaps = 10/290 (3%)
Frame = +1
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVR-------RIAKTEG 67
F + L GG++ +KT AP+ER+K+L Q + ++ LS + R K EG
Sbjct: 454 FFIDFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKTGRLSEPYKGIGDCFARTTKDEG 633
Query: 68 LLGFYRGNGASVARIIPYAGLHFMSYEEYRRL--IMQAFPNVWKGPTLDLMAGSLSGGTA 125
++ +RGN A+V R P L+F + ++RL + WK +L +G +G ++
Sbjct: 634 IVALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKWFAGNLASGGAAGASS 813
Query: 126 VLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVA 185
+LF Y LD RT+LA + K E+ + G+ D KT G+ GLYRG
Sbjct: 814 LLFVYSLDYARTRLANDAKAAKK-------GGERQFNGLVDVYRKTLASDGVAGLYRGFN 972
Query: 186 PTLFGIFPYAGLKFYFYEEMKR-RVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQ 244
+ GI Y GL F Y+ +K + + S A G + +YP++ VRR+
Sbjct: 973 ISCVGIIVYRGLYFGMYDSLKPVLLTGKLQDSFFASFGLGWLITNGAGLASYPIDTVRRR 1152
Query: 245 MQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAI 294
M + + E + K ++ + I + +G K+LF G N ++ V A +
Sbjct: 1153MM---MTSGEAVKYKSSLDAFQQILKNEGAKSLFKGAGANILRAVAGAGV 1293
Score = 65.1 bits (157), Expect = 1e-11
Identities = 51/201 (25%), Positives = 89/201 (43%), Gaps = 7/201 (3%)
Frame = +1
Query: 113 LDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 172
+D + G +S + P++ R KL Q + ++ +G ++ + Y+GI DC ++T
Sbjct: 460 IDFLMGGVSAAVSKTAAAPIE--RVKLLIQ--NQDEMIKTGRLS--EPYKGIGDCFARTT 621
Query: 173 KEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKR-----RVPEDYKKSIMAKLTCGSVA 227
K+ GI L+RG + FP L F F + KR + + Y K L G A
Sbjct: 622 KDEGIVALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKWFAGNLASGGAA 801
Query: 228 GLLGQTFTYPLEVVRRQMQVQNLAASE--EAELKGTMRSMVLIAQKQGWKTLFSGLSINY 285
G F Y L+ R ++ AA + E + G + G L+ G +I+
Sbjct: 802 GASSLLFVYSLDYARTRLANDAKAAKKGGERQFNGLVDVYRKTLASDGVAGLYRGFNISC 981
Query: 286 IKVVPSAAIGFTVYDTMKSYL 306
+ ++ + F +YD++K L
Sbjct: 982 VGIIVYRGLYFGMYDSLKPVL 1044
>TC18745 weakly similar to UP|ADT2_HUMAN (P05141) ADP,ATP carrier protein,
fibroblast isoform (ADP/ATP translocase 2) (Adenine
nucleotide translocator 2) (ANT 2), partial (9%)
Length = 834
Score = 114 bits (284), Expect = 3e-26
Identities = 69/196 (35%), Positives = 97/196 (49%), Gaps = 10/196 (5%)
Frame = +3
Query: 18 ELLAGGLAGGFAKTVVAPLERLKILFQTRR-----TEFRSAGLSGSVRRIAKTEGLLGFY 72
+LLAGG+AG F+KT APL RL ILFQ + + R + RI EG F
Sbjct: 282 QLLAGGVAGAFSKTCTAPLARLTILFQVQGMHSDFSALRKPSIWQEASRIINEEGFRAFG 461
Query: 73 RGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPT-----LDLMAGSLSGGTAVL 127
+GN ++ +PY+ + F SYE Y+ L ++G + + M G L+G T+
Sbjct: 462 KGNMVTIVHRLPYSAVSFYSYERYKNLFHSLMGENYRGSSSANLFVHFMGGGLAGVTSAS 641
Query: 128 FTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPT 187
TYPLDL+RT+LA Q + YRGI S ++ G GLY+G+ T
Sbjct: 642 ATYPLDLVRTRLAAQ-------------RSTIYYRGISHAFSTICRDEGFLGLYKGLGAT 782
Query: 188 LFGIFPYAGLKFYFYE 203
L G+ P + F YE
Sbjct: 783 LLGVGPSIAISFSVYE 830
Score = 67.0 bits (162), Expect = 4e-12
Identities = 57/202 (28%), Positives = 90/202 (44%), Gaps = 10/202 (4%)
Frame = +3
Query: 110 GPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYR--GIRDC 167
G L+AG ++G + T PL R + +Q V GM ++ R I
Sbjct: 270 GTVNQLLAGGVAGAFSKTCTAPL--ARLTILFQ--------VQGMHSDFSALRKPSIWQE 419
Query: 168 LSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKR----RVPEDYKKSIMAKLTC 223
S+ E G R +G T+ PY+ + FY YE K + E+Y+ S A L
Sbjct: 420 ASRIINEEGFRAFGKGNMVTIVHRLPYSAVSFYSYERYKNLFHSLMGENYRGSSSANLFV 599
Query: 224 ----GSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFS 279
G +AG+ + TYPL++VR ++ Q +G + I + +G+ L+
Sbjct: 600 HFMGGGLAGVTSASATYPLDLVRTRLAAQR----STIYYRGISHAFSTICRDEGFLGLYK 767
Query: 280 GLSINYIKVVPSAAIGFTVYDT 301
GL + V PS AI F+VY++
Sbjct: 768 GLGATLLGVGPSIAISFSVYES 833
>TC7883 homologue to UP|ADT1_GOSHI (O22342) ADP,ATP carrier protein 1,
mitochondrial precursor (ADP/ATP translocase 1) (Adenine
nucleotide translocator 1) (ANT 1), partial (54%)
Length = 856
Score = 108 bits (269), Expect = 2e-24
Identities = 67/201 (33%), Positives = 100/201 (49%), Gaps = 9/201 (4%)
Frame = +2
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSA-------GLSGSVRRIAKTEG 67
F + L GG++ +KT AP+ER+K+L Q + +S G+S R K EG
Sbjct: 272 FLADFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKSGRLSEPYKGISDCFGRTMKDEG 451
Query: 68 LLGFYRGNGASVARIIPYAGLHFMSYEEYRRL--IMQAFPNVWKGPTLDLMAGSLSGGTA 125
++ +RGN A+V R P L+F + ++RL + WK +L +G +G ++
Sbjct: 452 VIALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAAGASS 631
Query: 126 VLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVA 185
+LF Y LD RT+LA + K E+ + G+ D KT K GI GLYRG
Sbjct: 632 LLFVYSLDYARTRLANDAKAAKK-------GGERQFNGLIDVYKKTIKSDGIAGLYRGFN 790
Query: 186 PTLFGIFPYAGLKFYFYEEMK 206
+ GI Y GL F Y+ +K
Sbjct: 791 ISCVGIIVYRGLYFGMYDSLK 853
Score = 66.2 bits (160), Expect = 7e-12
Identities = 50/197 (25%), Positives = 87/197 (43%), Gaps = 7/197 (3%)
Frame = +2
Query: 114 DLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK 173
D + G +S + P++ R KL Q + ++ SG ++ + Y+GI DC +T K
Sbjct: 281 DFLMGGVSAAVSKTAAAPIE--RVKLLIQ--NQDEMIKSGRLS--EPYKGISDCFGRTMK 442
Query: 174 EGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKR-----RVPEDYKKSIMAKLTCGSVAG 228
+ G+ L+RG + FP L F F + KR + + Y K L G AG
Sbjct: 443 DEGVIALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAAG 622
Query: 229 LLGQTFTYPLEVVRRQMQVQNLAASE--EAELKGTMRSMVLIAQKQGWKTLFSGLSINYI 286
F Y L+ R ++ AA + E + G + + G L+ G +I+ +
Sbjct: 623 ASSLLFVYSLDYARTRLANDAKAAKKGGERQFNGLIDVYKKTIKSDGIAGLYRGFNISCV 802
Query: 287 KVVPSAAIGFTVYDTMK 303
++ + F +YD++K
Sbjct: 803 GIIVYRGLYFGMYDSLK 853
>TC15025 similar to UP|MCAT_ARATH (Q93XM7) Mitochondrial
carnitine/acylcarnitine carrier-like protein (A BOUT DE
SOUFFLE) (Carnitine/acylcarnitine translocase-like
protein) (CAC-like protein), partial (97%)
Length = 1256
Score = 88.6 bits (218), Expect = 1e-18
Identities = 73/300 (24%), Positives = 135/300 (44%), Gaps = 9/300 (3%)
Frame = +2
Query: 16 AKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEF-----RSAGLSGSVRRIAKTEGLLG 70
AK+L AG + G V P + +K+ Q++ T + +G +V++ EG G
Sbjct: 173 AKDLAAGTVGGASQLIVGHPFDTIKVKLQSQPTPLPGQPPKFSGAFDAVKQTIAAEGPRG 352
Query: 71 FYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTY 130
++G GA +A + + + F + I+++ P + + G+ +G
Sbjct: 353 LFKGMGAPLATVAAFNAVLF-TVRGQMETIVRSHPGAPLTVSQQFICGAGAGVAVSFLAC 529
Query: 131 PLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK-EGGIRGLYRGVAPTLF 189
P +LI+ +L Q + L SG Y G D + + EGG RGL++G+ PT+
Sbjct: 530 PTELIKCRLQAQ----SALGGSGAAAVAVKYGGPMDVARQVLRSEGGTRGLFKGLVPTMA 697
Query: 190 GIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQTF---TYPLEVVRRQMQ 246
P + F YE +K+++ S + + + GL G +F YP +VV+ +Q
Sbjct: 698 REIPGNAIMFGVYEAIKQQIAGGTDTSGLGRGSLIVAGGLAGASFWFLVYPTDVVKSVIQ 877
Query: 247 VQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
V + + + G++ + I +G+K L+ G + +P+ A F Y+ +S L
Sbjct: 878 VDDY---KNPKFSGSLDAFRKIQATEGFKGLYKGFGPAMARSIPANAACFLAYEMTRSAL 1048
>AV408895
Length = 429
Score = 88.6 bits (218), Expect = 1e-18
Identities = 52/152 (34%), Positives = 76/152 (49%), Gaps = 6/152 (3%)
Frame = +3
Query: 61 RIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAF------PNVWKGPTLD 114
RI EG+ F++GN ++A +PY+ ++F SYE Y++ + NV +
Sbjct: 12 RIVNEEGVRAFWKGNLVTIAHRLPYSSVNFYSYEHYKKWLRMVSGVQNHRDNVSADVFIH 191
Query: 115 LMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 174
+ G ++G TA TYPLDL+RT+LA Q N YRGI L KE
Sbjct: 192 FVGGGMAGITAATSTYPLDLVRTRLAAQ-------------TNFTYYRGIWHALQTISKE 332
Query: 175 GGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMK 206
GI GLY+G+ TL + P + F YE ++
Sbjct: 333 EGILGLYKGLGTTLLTVGPNIAISFSVYESLR 428
Score = 65.1 bits (157), Expect = 1e-11
Identities = 37/144 (25%), Positives = 71/144 (48%), Gaps = 9/144 (6%)
Frame = +3
Query: 169 SKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRV---------PEDYKKSIMA 219
S+ E G+R ++G T+ PY+ + FY YE K+ + ++ +
Sbjct: 9 SRIVNEEGVRAFWKGNLVTIAHRLPYSSVNFYSYEHYKKWLRMVSGVQNHRDNVSADVFI 188
Query: 220 KLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFS 279
G +AG+ T TYPL++VR ++ Q + +G ++ I++++G L+
Sbjct: 189 HFVGGGMAGITAATSTYPLDLVRTRLAAQ----TNFTYYRGIWHALQTISKEEGILGLYK 356
Query: 280 GLSINYIKVVPSAAIGFTVYDTMK 303
GL + V P+ AI F+VY++++
Sbjct: 357 GLGTTLLTVGPNIAISFSVYESLR 428
Score = 47.4 bits (111), Expect = 3e-06
Identities = 28/80 (35%), Positives = 45/80 (56%), Gaps = 2/80 (2%)
Frame = +3
Query: 20 LAGGLAGGFAKTVVAPLE--RLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNGA 77
+ GG+AG A T PL+ R ++ QT T +R G+ +++ I+K EG+LG Y+G G
Sbjct: 195 VGGGMAGITAATSTYPLDLVRTRLAAQTNFTYYR--GIWHALQTISKEEGILGLYKGLGT 368
Query: 78 SVARIIPYAGLHFMSYEEYR 97
++ + P + F YE R
Sbjct: 369 TLLTVGPNIAISFSVYESLR 428
>TC9482 similar to UP|Q9FI73 (Q9FI73) Mitochondrial carrier protein-like
(At5g48970), partial (42%)
Length = 529
Score = 72.0 bits (175), Expect = 1e-13
Identities = 41/149 (27%), Positives = 68/149 (45%), Gaps = 18/149 (12%)
Frame = +3
Query: 176 GIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKR-----------RVPEDYKKSIMAKLTCG 224
G +G+Y G++PTL I PYAGL+F Y+ KR + S CG
Sbjct: 9 GFQGMYAGLSPTLVEIIPYAGLQFGTYDTFKRWAMAWNHYRYSNTSAENSPSSFQLFLCG 188
Query: 225 SVAGLLGQTFTYPLEVVRRQMQVQNL-------AASEEAELKGTMRSMVLIAQKQGWKTL 277
AG + +PL+VV+++ Q++ L A E ++ I + +GW L
Sbjct: 189 LAAGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRAYSSMFDAIQRILRLEGWAGL 368
Query: 278 FSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
+ G+ + +K P+ A+ F Y+ +L
Sbjct: 369 YKGIIPSTVKAAPAGAVTFVAYELTSDWL 455
Score = 57.8 bits (138), Expect = 2e-09
Identities = 38/147 (25%), Positives = 65/147 (43%), Gaps = 8/147 (5%)
Frame = +3
Query: 65 TEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLD--------LM 116
T G G Y G ++ IIPYAGL F +Y+ ++R M + + + +
Sbjct: 3 TRGFQGMYAGLSPTLVEIIPYAGLQFGTYDTFKRWAMAWNHYRYSNTSAENSPSSFQLFL 182
Query: 117 AGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 176
G +G A L +PLD+++ + +QI + G + Y + D + + + G
Sbjct: 183 CGLAAGTCAKLVCHPLDVVKKR--FQIEGLQRHPRYGARVERRAYSSMFDAIQRILRLEG 356
Query: 177 IRGLYRGVAPTLFGIFPYAGLKFYFYE 203
GLY+G+ P+ P + F YE
Sbjct: 357 WAGLYKGIIPSTVKAAPAGAVTFVAYE 437
Score = 37.7 bits (86), Expect = 0.003
Identities = 29/101 (28%), Positives = 47/101 (45%), Gaps = 11/101 (10%)
Frame = +3
Query: 5 SESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQTR----------RTEFRS-A 53
S + ++ P + L G AG AK V PL+ +K FQ R E R+ +
Sbjct: 135 SNTSAENSPSSFQLFLCGLAAGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRAYS 314
Query: 54 GLSGSVRRIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYE 94
+ +++RI + EG G Y+G S + P + F++YE
Sbjct: 315 SMFDAIQRILRLEGWAGLYKGIIPSTVKAAPAGAVTFVAYE 437
Score = 31.6 bits (70), Expect = 0.18
Identities = 11/34 (32%), Positives = 23/34 (67%)
Frame = +3
Query: 272 QGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSY 305
+G++ +++GLS ++++P A + F YDT K +
Sbjct: 6 RGFQGMYAGLSPTLVEIIPYAGLQFGTYDTFKRW 107
>BP039065
Length = 564
Score = 68.6 bits (166), Expect = 1e-12
Identities = 36/100 (36%), Positives = 56/100 (56%), Gaps = 7/100 (7%)
Frame = +3
Query: 7 SILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEF-------RSAGLSGSV 59
+IL +P A AG +AG AKTV APL+R+K+L QT + G ++
Sbjct: 234 AILAFVPRDASLFSAGAIAGAAAKTVTAPLDRIKLLMQTHGVRVGQDSAKKAAIGFVEAI 413
Query: 60 RRIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRL 99
I K EG+ G+++GN V R+IPY+ + +YE Y+++
Sbjct: 414 TVIGKEEGIRGYWKGNLPQVIRVIPYSAVQLFAYEIYKKI 533
Score = 41.6 bits (96), Expect = 2e-04
Identities = 25/91 (27%), Positives = 45/91 (48%)
Frame = +3
Query: 117 AGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 176
AG+++G A T PLD I+ + V G + ++ G + ++ KE G
Sbjct: 276 AGAIAGAAAKTVTAPLDRIKLLMQTHGVRV------GQDSAKKAAIGFVEAITVIGKEEG 437
Query: 177 IRGLYRGVAPTLFGIFPYAGLKFYFYEEMKR 207
IRG ++G P + + PY+ ++ + YE K+
Sbjct: 438 IRGYWKGNLPQVIRVIPYSAVQLFAYEIYKK 530
Score = 41.2 bits (95), Expect = 2e-04
Identities = 21/89 (23%), Positives = 46/89 (51%), Gaps = 3/89 (3%)
Frame = +3
Query: 222 TCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELK---GTMRSMVLIAQKQGWKTLF 278
+ G++AG +T T PL+ ++ MQ + +++ K G + ++ +I +++G + +
Sbjct: 273 SAGAIAGAAAKTVTAPLDRIKLLMQTHGVRVGQDSAKKAAIGFVEAITVIGKEEGIRGYW 452
Query: 279 SGLSINYIKVVPSAAIGFTVYDTMKSYLR 307
G I+V+P +A+ Y+ K R
Sbjct: 453 KGNLPQVIRVIPYSAVQLFAYEIYKKIFR 539
>CB827990
Length = 547
Score = 67.0 bits (162), Expect = 4e-12
Identities = 48/160 (30%), Positives = 76/160 (47%), Gaps = 22/160 (13%)
Frame = +3
Query: 173 KEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRV---PEDYKKSIMAK--------- 220
K G LY+G+ P++ + P + + Y+ +K PE K+ K
Sbjct: 9 KTEGFFSLYKGLVPSIVSMAPSGAVFYGVYDILKSAYLHSPEGMKRIQHMKEETEELNAF 188
Query: 221 ----------LTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQ 270
L G++AG + TYP EVVRRQ+Q+Q A T+ + V I +
Sbjct: 189 DQLELGPVRTLLYGAIAGCCAEAATYPFEVVRRQLQMQ-----VRATRLNTLATCVKIVE 353
Query: 271 KQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRVPS 310
+ G ++GL + ++V+PSAAI + VY+ MK L+V S
Sbjct: 354 QGGVPAFYAGLIPSLLQVLPSAAISYFVYEFMKIVLKVES 473
Score = 59.3 bits (142), Expect = 8e-10
Identities = 41/162 (25%), Positives = 70/162 (42%), Gaps = 19/162 (11%)
Frame = +3
Query: 64 KTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQ-------------------AF 104
KTEG Y+G S+ + P + + Y+ + + AF
Sbjct: 9 KTEGFFSLYKGLVPSIVSMAPSGAVFYGVYDILKSAYLHSPEGMKRIQHMKEETEELNAF 188
Query: 105 PNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGI 164
+ GP L+ G+++G A TYP +++R +L Q V T+LN +
Sbjct: 189 DQLELGPVRTLLYGAIAGCCAEAATYPFEVVRRQLQMQ-VRATRLNT------------L 329
Query: 165 RDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMK 206
C+ K ++GG+ Y G+ P+L + P A + ++ YE MK
Sbjct: 330 ATCV-KIVEQGGVPAFYAGLIPSLLQVLPSAAISYFVYEFMK 452
>TC7843 weakly similar to GB|AAF03412.1|6090963|AF188712 mitochondrial
dicarboxylate carrier {Mus musculus;} , partial (22%)
Length = 1594
Score = 65.5 bits (158), Expect = 1e-11
Identities = 53/210 (25%), Positives = 100/210 (47%), Gaps = 7/210 (3%)
Frame = +3
Query: 11 HIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQT-------RRTEFRSAGLSGSVRRIA 63
++PL ++++ AG +AGG V P + + Q +R + S + ++ R+A
Sbjct: 630 NMPL-SRKIEAGLIAGGIGAAVGNPADVAMVRMQADGRLPAAQRRNYSS--VVDAITRMA 800
Query: 64 KTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGG 123
K EG+ +RG+ +V R + SY++++ +I++ + G + A +G
Sbjct: 801 KQEGVTSLWRGSSLTVNRAMLVTASQLASYDQFKEMILEK-GLMRDGLGTHVTASFAAGF 977
Query: 124 TAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRG 183
A + + P+D+I+T++ V P E Y G DC KT + G LY+G
Sbjct: 978 VAAVASNPVDVIKTRVMNMKVEP---------GAEPPYAGALDCALKTVRAEGPMALYKG 1130
Query: 184 VAPTLFGIFPYAGLKFYFYEEMKRRVPEDY 213
PT+ P+ + F E++ R++ +DY
Sbjct: 1131FIPTISRQGPFTVVLFVTLEQV-RKMFKDY 1217
>TC10512 similar to UP|Q9FI73 (Q9FI73) Mitochondrial carrier protein-like
(At5g48970), partial (43%)
Length = 563
Score = 63.5 bits (153), Expect = 4e-11
Identities = 38/136 (27%), Positives = 68/136 (49%), Gaps = 19/136 (13%)
Frame = +1
Query: 21 AGGLAGGFAKTVVAPLERLKILFQTRRTEFRS----------------AGLSGSVRRIAK 64
AG ++GG ++TV +PL+ +KI FQ + S G+ + + I +
Sbjct: 151 AGAISGGISRTVTSPLDVIKIRFQVQLEPTSSWALLRRNLAATAPSKYTGMFQASKDILR 330
Query: 65 TEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLI---MQAFPNVWKGPTLDLMAGSLS 121
EG+ GF+RGN ++ ++PY + F + + + ++ P L ++G+L+
Sbjct: 331 EEGVQGFWRGNVPALLMVMPYTAIQFTVLHKLKTFASGSSKTENHIGLSPYLSYVSGALA 510
Query: 122 GGTAVLFTYPLDLIRT 137
G A L +YP DL+RT
Sbjct: 511 GCAATLGSYPFDLLRT 558
Score = 53.9 bits (128), Expect = 3e-08
Identities = 38/148 (25%), Positives = 64/148 (42%), Gaps = 10/148 (6%)
Frame = +1
Query: 105 PNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTK----LNVSGMVNNEQV 160
P+ K +D AG++SGG + T PLD+I+ + Q+ PT L +
Sbjct: 115 PSQLKRAMIDSSAGAISGGISRTVTSPLDVIKIRFQVQL-EPTSSWALLRRNLAATAPSK 291
Query: 161 YRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKK----- 215
Y G+ +E G++G +RG P L + PY ++F ++K K
Sbjct: 292 YTGMFQASKDILREEGVQGFWRGNVPALLMVMPYTAIQFTVLHKLKTFASGSSKTENHIG 471
Query: 216 -SIMAKLTCGSVAGLLGQTFTYPLEVVR 242
S G++AG +YP +++R
Sbjct: 472 LSPYLSYVSGALAGCAATLGSYPFDLLR 555
Score = 44.7 bits (104), Expect = 2e-05
Identities = 30/111 (27%), Positives = 56/111 (50%), Gaps = 12/111 (10%)
Frame = +1
Query: 215 KSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQ------------NLAASEEAELKGTM 262
K M + G+++G + +T T PL+V++ + QVQ NLAA+ ++ G
Sbjct: 127 KRAMIDSSAGAISGGISRTVTSPLDVIKIRFQVQLEPTSSWALLRRNLAATAPSKYTGMF 306
Query: 263 RSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDE 313
++ I +++G + + G + V+P AI FTV +K++ S+ E
Sbjct: 307 QASKDILREEGVQGFWRGNVPALLMVMPYTAIQFTVLHKLKTFASGSSKTE 459
>TC9406 similar to UP|Q9M058 (Q9M058) Ca-dependent solute carrier-like
protein, partial (30%)
Length = 676
Score = 62.8 bits (151), Expect = 7e-11
Identities = 35/114 (30%), Positives = 58/114 (50%), Gaps = 4/114 (3%)
Frame = +3
Query: 197 LKFYFYEEMKR----RVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAA 252
+ F YE ++ R P+D S + L C S++G+ T +PL++VRR+MQ++ +
Sbjct: 12 ISFSVYESLRSLWISRRPDD--SSAVVSLACASLSGVASSTVIFPLDLVRRRMQLEGVGG 185
Query: 253 SEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
G + I + +G + L+ G+ Y KVVP I F Y+T+K L
Sbjct: 186 RACVYNTGMCGAFQCIIKNEGVRGLYRGILPEYYKVVPGVGIAFMTYETLKRLL 347
Score = 53.9 bits (128), Expect = 3e-08
Identities = 36/121 (29%), Positives = 54/121 (43%), Gaps = 1/121 (0%)
Frame = +3
Query: 88 LHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPT 147
+ F YE R L + P+ + L SLSG + +PLDL+R ++
Sbjct: 12 ISFSVYESLRSLWISRRPDD-SSAVVSLACASLSGVASSTVIFPLDLVRRRM-------- 164
Query: 148 KLNVSGMVNNEQVYR-GIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMK 206
+ G+ VY G+ K G+RGLYRG+ P + + P G+ F YE +K
Sbjct: 165 --QLEGVGGRACVYNTGMCGAFQCIIKNEGVRGLYRGILPEYYKVVPGVGIAFMTYETLK 338
Query: 207 R 207
R
Sbjct: 339 R 341
Score = 47.8 bits (112), Expect = 2e-06
Identities = 27/86 (31%), Positives = 45/86 (51%), Gaps = 4/86 (4%)
Frame = +3
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSA----GLSGSVRRIAKTEGLLGFYRG 74
L L+G + TV+ PL+ ++ Q R+ G+ G+ + I K EG+ G YRG
Sbjct: 90 LACASLSGVASSTVIFPLDLVRRRMQLEGVGGRACVYNTGMCGAFQCIIKNEGVRGLYRG 269
Query: 75 NGASVARIIPYAGLHFMSYEEYRRLI 100
+++P G+ FM+YE +RL+
Sbjct: 270 ILPEYYKVVPGVGIAFMTYETLKRLL 347
>BP071178
Length = 408
Score = 60.5 bits (145), Expect = 4e-10
Identities = 34/112 (30%), Positives = 61/112 (54%)
Frame = +2
Query: 106 NVWKGPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIR 165
N++ G + + +AG+ +G T+++ YPLD+ T+LA I G Q +RGI
Sbjct: 89 NLFSGASANFVAGAAAGCTSLVLVYPLDIAHTRLAADI---------GRTEVRQ-FRGIH 238
Query: 166 DCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSI 217
L + + GIRG+YRG+ +L G+ + GL F ++ +K + E+ + +
Sbjct: 239 HFLVTIFHKDGIRGIYRGLPASLHGMVVHRGLYFGGFDTIKEMLTEESEPEV 394
>TC8441 homologue to UP|ADT2_SOLTU (P27081) ADP,ATP carrier protein,
mitochondrial precursor (ADP/ATP translocase) (Adenine
nucleotide translocator) (ANT) (Fragment), partial (48%)
Length = 884
Score = 59.3 bits (142), Expect = 8e-10
Identities = 37/129 (28%), Positives = 60/129 (45%), Gaps = 1/129 (0%)
Frame = +1
Query: 167 CLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKR-RVPEDYKKSIMAKLTCGS 225
CL + G+ GLYRG + GI Y GL F Y+ +K + + S A G
Sbjct: 130 CLQEDLASDGVAGLYRGFNISCVGIIVYRGLYFGMYDSLKPVLLTGSLQDSFFASFGLGW 309
Query: 226 VAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINY 285
+ +YP++ VRR+M + + E + K +M + I + +G K+LF G N
Sbjct: 310 LITNGAGLASYPIDTVRRRMM---MTSGEAVKYKSSMDAFTQILKNEGPKSLFKGAGANI 480
Query: 286 IKVVPSAAI 294
++ V A +
Sbjct: 481 LRAVAGAGV 507
>BP062301
Length = 364
Score = 58.5 bits (140), Expect = 1e-09
Identities = 29/77 (37%), Positives = 47/77 (60%), Gaps = 1/77 (1%)
Frame = +1
Query: 29 AKTVVAPLERLKILFQTRRTE-FRSAGLSGSVRRIAKTEGLLGFYRGNGASVARIIPYAG 87
++T VAPLER+KIL Q + + G ++ I +TEG G ++GNG + ARI+P +
Sbjct: 106 SRTAVAPLERMKILLQVQNPHNIKYNGTIQGLKYIWRTEGFRGLFKGNGTNCARIVPNSA 285
Query: 88 LHFMSYEEYRRLIMQAF 104
+ F SYE+ + I+ +
Sbjct: 286 VKFFSYEQASKGILHLY 336
Score = 34.3 bits (77), Expect = 0.028
Identities = 19/82 (23%), Positives = 37/82 (44%)
Frame = +1
Query: 134 LIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFP 193
L R K+ Q+ +P + +G + L ++ G RGL++G I P
Sbjct: 127 LERMKILLQVQNPHNIKYNGTIQG----------LKYIWRTEGFRGLFKGNGTNCARIVP 276
Query: 194 YAGLKFYFYEEMKRRVPEDYKK 215
+ +KF+ YE+ + + Y++
Sbjct: 277 NSAVKFFSYEQASKGILHLYRQ 342
>TC15127 weakly similar to GB|AAG29586.1|11094343|AF155813 mitochondrial
uncoupling protein 5 short form {Mus musculus;}, partial
(24%)
Length = 650
Score = 52.8 bits (125), Expect = 8e-08
Identities = 37/147 (25%), Positives = 69/147 (46%), Gaps = 1/147 (0%)
Frame = +3
Query: 66 EGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIM-QAFPNVWKGPTLDLMAGSLSGGT 124
EG+ +RG+G +V R + SY++++ I+ + + N G + A +G
Sbjct: 9 EGIGSLWRGSGLTVNRAMIVTASQLASYDQFKETIVGKGWMN--DGIGTHVAASFAAGFV 182
Query: 125 AVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGV 184
A + + P+D+I+T++ +N+ Y G DC KT K G LY+G
Sbjct: 183 AAVASNPVDVIKTRV---------MNMKVDAGEAPPYNGALDCALKTVKAEGPLALYKGF 335
Query: 185 APTLFGIFPYAGLKFYFYEEMKRRVPE 211
PT+ P+ + F E++++ + E
Sbjct: 336 IPTISRQGPFTVVLFVTLEQVRKLLKE 416
>TC8324 UP|Q8VWY1 (Q8VWY1) Mitochondrial phosphate transporter, complete
Length = 1605
Score = 52.4 bits (124), Expect = 1e-07
Identities = 47/193 (24%), Positives = 79/193 (40%), Gaps = 6/193 (3%)
Frame = +3
Query: 116 MAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEG 175
+ G LS G + PLDL++ + + PTK Y+ I KE
Sbjct: 342 VGGILSCGLTHMTVTPLDLVKCNMQ---IDPTK------------YKSISSGFGVLLKEQ 476
Query: 176 GIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPE----DY--KKSIMAKLTCGSVAGL 229
G++G +RG PTL G KF FYE K+ + +Y K + L + A +
Sbjct: 477 GVKGFFRGWVPTLLGYSAQGACKFGFYEFFKKYYSDIAGPEYATKYKTLIYLAGSASAEV 656
Query: 230 LGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVV 289
+ P E V+ ++Q Q A +G + + +G L+ GL + + +
Sbjct: 657 IADVALCPFEAVKVRVQTQPGFA------RGLSDGLPKFVKAEGTLGLYKGLVPLWGRQI 818
Query: 290 PSAAIGFTVYDTM 302
P + F ++T+
Sbjct: 819 PYTMMKFASFETI 857
Score = 42.0 bits (97), Expect = 1e-04
Identities = 55/192 (28%), Positives = 82/192 (42%), Gaps = 9/192 (4%)
Frame = +3
Query: 24 LAGGFAKTVVA-----PLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNGAS 78
LAG + V+A P E +K+ QT+ R GLS + + K EG LG Y+G
Sbjct: 630 LAGSASAEVIADVALCPFEAVKVRVQTQPGFAR--GLSDGLPKFVKAEGTLGLYKGLVPL 803
Query: 79 VARIIPYAGLHFMSYEEYRRLIMQ-AFPNVWKGPTLDLMAG-SLSGG-TAVLFTYPLDLI 135
R IPY + F S+E +I + A P + +L G S +GG A +F
Sbjct: 804 WGRQIPYTMMKFASFETIVEMIYKHAVPVPKSECSKNLQLGISFAGGYVAGVFC------ 965
Query: 136 RTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLY-RGVAPTLFGIFPY 194
IVS N+ +NN Q + K+ G+ GL+ RG+ + I
Sbjct: 966 ------AIVSHPADNLVSFLNNAQ-----GATVGDAVKKLGMWGLFTRGLPLRIVMIGTL 1112
Query: 195 AGLKFYFYEEMK 206
G ++ Y+ K
Sbjct: 1113TGAQWGIYDAFK 1148
>TC13453 similar to UP|Q9ZNY4 (Q9ZNY4) Mitochondrial energy transfer protein
precursor, partial (18%)
Length = 506
Score = 50.4 bits (119), Expect = 4e-07
Identities = 23/60 (38%), Positives = 35/60 (58%)
Frame = +2
Query: 153 GMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPED 212
G + QVY+ + L+ ++ GI+GLYRG+ P+ + P AG+ F YE KR + ED
Sbjct: 35 GAXSGRQVYKNVLHALASILEKEGIQGLYRGLGPSCMKLVPAAGISFMCYEACKRILVED 214
Score = 46.2 bits (108), Expect = 7e-06
Identities = 26/70 (37%), Positives = 39/70 (55%)
Frame = +2
Query: 237 PLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGF 296
PLEV R+ MQV A S K + ++ I +K+G + L+ GL + +K+VP+A I F
Sbjct: 2 PLEVARKHMQVG--AXSGRQVYKNVLHALASILEKEGIQGLYRGLGPSCMKLVPAAGISF 175
Query: 297 TVYDTMKSYL 306
Y+ K L
Sbjct: 176 MCYEACKRIL 205
Score = 42.4 bits (98), Expect = 1e-04
Identities = 17/41 (41%), Positives = 28/41 (67%)
Frame = +2
Query: 62 IAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQ 102
I + EG+ G YRG G S +++P AG+ FM YE +R++++
Sbjct: 89 ILEKEGIQGLYRGLGPSCMKLVPAAGISFMCYEACKRILVE 211
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.139 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,172,665
Number of Sequences: 28460
Number of extensions: 45680
Number of successful extensions: 303
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 230
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 277
length of query: 315
length of database: 4,897,600
effective HSP length: 90
effective length of query: 225
effective length of database: 2,336,200
effective search space: 525645000
effective search space used: 525645000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC148360.5


