

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148360.2 + phase: 0 /pseudo
(322 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
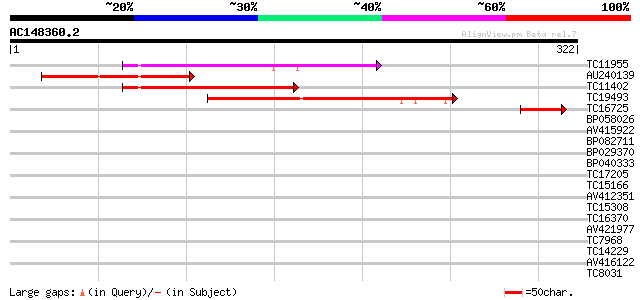
Score E
Sequences producing significant alignments: (bits) Value
TC11955 similar to UP|Q9LHH9 (Q9LHH9) Protein arginine N-methylt... 131 1e-31
AU240139 130 2e-31
TC11402 similar to UP|Q9LJZ9 (Q9LJZ9) Protein arginine N-methylt... 110 4e-25
TC19493 similar to GB|AAH36974.1|22477705|BC036974 Carm1-pending... 102 1e-22
TC16725 similar to UP|Q9SU94 (Q9SU94) Arginine methyltransferase... 53 8e-08
BP058026 38 0.002
AV415922 38 0.003
BP082711 37 0.006
BP029370 33 0.085
BP040333 32 0.14
TC17205 weakly similar to PIR|S50830|S50830 Machado-Joseph disea... 30 0.42
TC15166 similar to GB|AAK28312.1|13506739|AF224702 WRKY DNA-bind... 30 0.55
AV412351 29 1.2
TC15308 similar to UP|Q41725 (Q41725) TED3, partial (16%) 29 1.2
TC16370 similar to UP|MOT3_YEAST (P54785) Zinc finger protein MO... 28 2.1
AV421977 28 2.1
TC7968 similar to UP|Q39360 (Q39360) (Clone PIS63-1), partial (15%) 28 2.7
TC14229 similar to UP|P93166 (P93166) SCOF-1, partial (38%) 28 2.7
AV416122 28 2.7
TC8031 similar to UP|Q9FR33 (Q9FR33) Ripening regulated protein ... 23 2.9
>TC11955 similar to UP|Q9LHH9 (Q9LHH9) Protein arginine N-methyltransferase
3-like protein, partial (24%)
Length = 572
Score = 131 bits (330), Expect = 1e-31
Identities = 78/165 (47%), Positives = 99/165 (59%), Gaps = 18/165 (10%)
Frame = +1
Query: 65 YFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGA 124
YF +YS FG H+EML D VR Y I +N L VV+DVG GTGILSLF A+AGA
Sbjct: 79 YFGAYSSFG-IHREMLSDKVRMDAYGQAILKNPSLLNGAVVMDVGCGTGILSLFAAQAGA 255
Query: 125 AHVYAVECS-HMADRAKEIVETNGY-------------SKVITVLKGKIEEL----ELPV 166
+ V AVE S MA A ++ + NG VI V+ +EE+ EL
Sbjct: 256 SRVIAVEASAKMAAVASQVAKDNGLWLSKSQSGVNGLQKGVIEVVNCMVEEIDKTVELQP 435
Query: 167 PKVDIIISEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASL 211
VD+++SEWMGY LL+E+ML SVL+ARD+WL G ILPD A++
Sbjct: 436 HSVDVLLSEWMGYCLLYESMLGSVLYARDRWLKPGGAILPDTATI 570
>AU240139
Length = 300
Score = 130 bits (328), Expect = 2e-31
Identities = 68/88 (77%), Positives = 77/88 (87%), Gaps = 1/88 (1%)
Frame = +1
Query: 19 NNNNNNHLRFEDAD-EVIEDVAETSNLDQSMGGCDLDDSNDKTSADYYFDSYSHFGNFHK 77
+ + HLRF+DAD EVIEDVAETSNLDQSM D DDS+DKTSADYYFDSYSHFG H+
Sbjct: 43 SQRQHEHLRFQDADDEVIEDVAETSNLDQSMCD-DPDDSDDKTSADYYFDSYSHFG-IHE 216
Query: 78 EMLKDTVRTKTYQNVIYQNRFLFKNKVV 105
EMLKDTVRTKTYQ+VIYQN+FLFK+K+V
Sbjct: 217 EMLKDTVRTKTYQSVIYQNKFLFKDKIV 300
>TC11402 similar to UP|Q9LJZ9 (Q9LJZ9) Protein arginine
N-methyltransferase-like protein, partial (28%)
Length = 536
Score = 110 bits (274), Expect = 4e-25
Identities = 53/100 (53%), Positives = 74/100 (74%)
Frame = +1
Query: 65 YFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGA 124
YF SY+H G H+EM+KD VRT+TY+ I +++ KVV+DVG GTGILS+FCA+AGA
Sbjct: 232 YFHSYAHLG-IHQEMIKDRVRTETYREAIMRHQSFIAGKVVVDVGCGTGILSIFCAQAGA 408
Query: 125 AHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELEL 164
VYAV+ S +A +A E+V+ N S VI VL G++E++E+
Sbjct: 409 KRVYAVDASDIALQANEVVKANNLSDVIVVLHGRVEDVEI 528
>TC19493 similar to GB|AAH36974.1|22477705|BC036974 Carm1-pending protein
{Mus musculus;} , partial (11%)
Length = 469
Score = 102 bits (253), Expect = 1e-22
Identities = 63/152 (41%), Positives = 92/152 (60%), Gaps = 10/152 (6%)
Frame = +3
Query: 113 GILSLFCAKAGAAHVYAVECSHMADRAKEIVETN-GYSKVITVLKGKIEELELPVPKVDI 171
GILSLF A+AGA HVYAVE S MA+ A++++ N ++ ITV+KGK+E++ELP K DI
Sbjct: 3 GILSLFAAQAGAKHVYAVEASEMAEYARKLIAGNPKLAQRITVIKGKVEDVELP-EKADI 179
Query: 172 IISEWMGYFLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDY---KEDKIEF 228
+ISE MG L+ E ML S + ARD++L G + P + +++ D+ +K F
Sbjct: 180 LISEPMGTLLVNERMLESYVIARDRFLTPTGKMFPAVGRIHMAPFTDEYLFIEVANKALF 359
Query: 229 W--NNVYGFDMSCIKKQALM----EPLVDTVD 254
W N YG D++ + A +P+VD D
Sbjct: 360 WQQQNYYGVDLTPLHGTAFQGYFSQPVVDAFD 455
>TC16725 similar to UP|Q9SU94 (Q9SU94) Arginine methyltransferase pam1
(PAM1) , partial (24%)
Length = 514
Score = 52.8 bits (125), Expect = 8e-08
Identities = 22/26 (84%), Positives = 24/26 (91%)
Frame = +3
Query: 291 DDFIHAFVAYFDVSFTKCHKLMGFST 316
DDFIHA +AYFDV+FT CHKLMGFST
Sbjct: 3 DDFIHALLAYFDVTFTMCHKLMGFST 80
>BP058026
Length = 389
Score = 38.1 bits (87), Expect = 0.002
Identities = 15/24 (62%), Positives = 17/24 (70%)
Frame = +3
Query: 3 SRKNNNQTNNTNMDNNNNNNNNHL 26
+ NNN NN N +NNNNNNNN L
Sbjct: 18 NNNNNNNNNNNNNNNNNNNNNNDL 89
Score = 37.7 bits (86), Expect = 0.003
Identities = 14/25 (56%), Positives = 18/25 (72%)
Frame = +3
Query: 1 MGSRKNNNQTNNTNMDNNNNNNNNH 25
+ + NNN NN N +NNNNNNNN+
Sbjct: 6 INNNNNNNNNNNNNNNNNNNNNNNN 80
Score = 37.4 bits (85), Expect = 0.003
Identities = 14/23 (60%), Positives = 17/23 (73%)
Frame = +3
Query: 3 SRKNNNQTNNTNMDNNNNNNNNH 25
+ NNN NN N +NNNNNNNN+
Sbjct: 15 NNNNNNNNNNNNNNNNNNNNNNN 83
Score = 32.7 bits (73), Expect = 0.085
Identities = 13/28 (46%), Positives = 19/28 (67%)
Frame = +3
Query: 8 NQTNNTNMDNNNNNNNNHLRFEDADEVI 35
N NN N +NNNNNNNN+ + +++I
Sbjct: 9 NNNNNNNNNNNNNNNNNNNNNNNNNDLI 92
>AV415922
Length = 422
Score = 37.7 bits (86), Expect = 0.003
Identities = 15/25 (60%), Positives = 18/25 (72%)
Frame = +1
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNHL 26
G NN+ NN N +NNNNNNNN+L
Sbjct: 163 GINTNNDDNNNNNNNNNNNNNNNNL 237
Score = 31.2 bits (69), Expect = 0.25
Identities = 11/23 (47%), Positives = 16/23 (68%)
Frame = +1
Query: 1 MGSRKNNNQTNNTNMDNNNNNNN 23
+ + ++N NN N +NNNNNNN
Sbjct: 166 INTNNDDNNNNNNNNNNNNNNNN 234
Score = 30.8 bits (68), Expect = 0.32
Identities = 12/20 (60%), Positives = 14/20 (70%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNH 25
N+ N N DNNNNNNNN+
Sbjct: 154 NSKGINTNNDDNNNNNNNNN 213
Score = 30.4 bits (67), Expect = 0.42
Identities = 12/21 (57%), Positives = 15/21 (71%)
Frame = +1
Query: 5 KNNNQTNNTNMDNNNNNNNNH 25
K N N+ N +NNNNNNNN+
Sbjct: 160 KGINTNNDDNNNNNNNNNNNN 222
>BP082711
Length = 379
Score = 36.6 bits (83), Expect = 0.006
Identities = 14/19 (73%), Positives = 15/19 (78%)
Frame = +3
Query: 6 NNNQTNNTNMDNNNNNNNN 24
NNN NN N +NNNNNNNN
Sbjct: 177 NNNNNNNNNNNNNNNNNNN 233
Score = 35.4 bits (80), Expect = 0.013
Identities = 13/21 (61%), Positives = 16/21 (75%)
Frame = +3
Query: 6 NNNQTNNTNMDNNNNNNNNHL 26
NNN NN N +NNNNNNN ++
Sbjct: 180 NNNNNNNNNNNNNNNNNNKNI 242
Score = 34.7 bits (78), Expect = 0.022
Identities = 13/19 (68%), Positives = 15/19 (78%)
Frame = +3
Query: 7 NNQTNNTNMDNNNNNNNNH 25
NN NN N +NNNNNNNN+
Sbjct: 177 NNNNNNNNNNNNNNNNNNN 233
Score = 32.3 bits (72), Expect = 0.11
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +3
Query: 3 SRKNNNQTNNTNMDNNNNNNN 23
+ NNN NN N +NNNNN N
Sbjct: 177 NNNNNNNNNNNNNNNNNNNKN 239
>BP029370
Length = 364
Score = 32.7 bits (73), Expect = 0.085
Identities = 20/63 (31%), Positives = 28/63 (43%), Gaps = 12/63 (19%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNN-----NNHLRFE-------DADEVIEDVAETSNLDQSMGGCDL 53
NNN NN N +NNNNN+ NH+ + DA + + T +L +G
Sbjct: 175 NNNNNNNKNNNNNNNNSII*KITNHITYASNFHHLWDASQQTHQFSHTLHLQGWLGSLAR 354
Query: 54 DDS 56
D S
Sbjct: 355 DSS 363
>BP040333
Length = 480
Score = 32.0 bits (71), Expect = 0.14
Identities = 13/23 (56%), Positives = 14/23 (60%)
Frame = +1
Query: 5 KNNNQTNNTNMDNNNNNNNNHLR 27
K NN N +NNNNNNN LR
Sbjct: 61 KTKTTNNNNNNNNNNNNNNKSLR 129
Score = 27.3 bits (59), Expect = 3.6
Identities = 15/39 (38%), Positives = 23/39 (58%)
Frame = +1
Query: 9 QTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQS 47
+T TN +NNNNNNNN+ + EV D ++ D++
Sbjct: 61 KTKTTN-NNNNNNNNNNNNNKSLREVS*DYEHSNIRDKT 174
Score = 26.2 bits (56), Expect = 7.9
Identities = 11/22 (50%), Positives = 14/22 (63%)
Frame = +1
Query: 3 SRKNNNQTNNTNMDNNNNNNNN 24
++ NN NN N NNNNNN +
Sbjct: 64 TKTTNNNNNNNN--NNNNNNKS 123
>TC17205 weakly similar to PIR|S50830|S50830 Machado-Joseph disease MJD1a
protein - human {Homo sapiens;}, partial (7%)
Length = 528
Score = 30.4 bits (67), Expect = 0.42
Identities = 13/35 (37%), Positives = 21/35 (59%)
Frame = +1
Query: 3 SRKNNNQTNNTNMDNNNNNNNNHLRFEDADEVIED 37
++ NNN +N TN +N+NN +++ D DE D
Sbjct: 310 AKTNNNNSNGTN-NNSNNKAQSNINLRDEDEFAAD 411
>TC15166 similar to GB|AAK28312.1|13506739|AF224702 WRKY DNA-binding protein
6 {Arabidopsis thaliana;} , partial (26%)
Length = 668
Score = 30.0 bits (66), Expect = 0.55
Identities = 14/23 (60%), Positives = 16/23 (68%)
Frame = +3
Query: 2 GSRKNNNQTNNTNMDNNNNNNNN 24
GS NNN ++N NNNNNNNN
Sbjct: 342 GSHANNNGSHN----NNNNNNNN 398
Score = 27.7 bits (60), Expect = 2.7
Identities = 10/18 (55%), Positives = 12/18 (66%)
Frame = +3
Query: 8 NQTNNTNMDNNNNNNNNH 25
+ NN NNNNNNNN+
Sbjct: 345 SHANNNGSHNNNNNNNNN 398
>AV412351
Length = 411
Score = 28.9 bits (63), Expect = 1.2
Identities = 24/92 (26%), Positives = 39/92 (42%)
Frame = +2
Query: 40 ETSNLDQSMGGCDLDDSNDKTSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFL 99
E L+ +M G + K+ Y +FHK + ++ + I Q +
Sbjct: 98 ERVRLESTMAGGKMKKEKGKSQQS---TPYQGGISFHKSKGQHILKNPLLVDAIVQKSGI 268
Query: 100 FKNKVVLDVGAGTGILSLFCAKAGAAHVYAVE 131
V+L++G GTG L+ +AG V AVE
Sbjct: 269 KSTDVILEIGPGTGNLTKKLLEAG-KKVIAVE 361
>TC15308 similar to UP|Q41725 (Q41725) TED3, partial (16%)
Length = 1103
Score = 28.9 bits (63), Expect = 1.2
Identities = 18/67 (26%), Positives = 31/67 (45%)
Frame = +3
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDKTS 61
G R N+N +NNNNN N+ +E E + D T ++ ++D N+ +
Sbjct: 498 GYRSNDNNGKELYYNNNNNNYGNNNGYERKREGMSD---TRFMENGKYHYNVDTENESYN 668
Query: 62 ADYYFDS 68
+ Y +S
Sbjct: 669 LNGYDES 689
>TC16370 similar to UP|MOT3_YEAST (P54785) Zinc finger protein MOT3/HMS1,
partial (4%)
Length = 545
Score = 28.1 bits (61), Expect = 2.1
Identities = 10/22 (45%), Positives = 17/22 (76%)
Frame = -3
Query: 6 NNNQTNNTNMDNNNNNNNNHLR 27
N+++ +N N DNNN+NN N+ +
Sbjct: 402 NHSRISNNNNDNNNSNNINNYK 337
>AV421977
Length = 535
Score = 28.1 bits (61), Expect = 2.1
Identities = 11/17 (64%), Positives = 12/17 (69%)
Frame = +3
Query: 8 NQTNNTNMDNNNNNNNN 24
N TN +NNNNNNNN
Sbjct: 12 NPTNTIIFNNNNNNNNN 62
Score = 26.6 bits (57), Expect = 6.1
Identities = 10/15 (66%), Positives = 13/15 (86%)
Frame = +3
Query: 12 NTNMDNNNNNNNNHL 26
NT + NNNNNNNN++
Sbjct: 21 NTIIFNNNNNNNNNI 65
>TC7968 similar to UP|Q39360 (Q39360) (Clone PIS63-1), partial (15%)
Length = 1193
Score = 27.7 bits (60), Expect = 2.7
Identities = 22/85 (25%), Positives = 38/85 (43%), Gaps = 12/85 (14%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNN--NNNH----------LRFEDADEVIEDVAETSNLDQSMGGCDL 53
NNN N N + NNNN NNN RF + + +V + N + G
Sbjct: 598 NNNAAANENYNFNNNNAANNNEKGAEVQGMSDTRFMEGGKYFYNV-NSENEKYNPTGYGG 774
Query: 54 DDSNDKTSADYYFDSYSHFGNFHKE 78
+ S +++ ++++ +FGN + E
Sbjct: 775 ESSTRGVNSENWYNNKGYFGNNNHE 849
>TC14229 similar to UP|P93166 (P93166) SCOF-1, partial (38%)
Length = 1175
Score = 27.7 bits (60), Expect = 2.7
Identities = 11/21 (52%), Positives = 14/21 (66%)
Frame = +1
Query: 11 NNTNMDNNNNNNNNHLRFEDA 31
NN N +NN +NN N LR E +
Sbjct: 259 NNNNKNNNTDNNTNTLRAESS 321
>AV416122
Length = 414
Score = 27.7 bits (60), Expect = 2.7
Identities = 20/70 (28%), Positives = 27/70 (38%), Gaps = 7/70 (10%)
Frame = +3
Query: 17 NNNNNNNNHLRFEDADEVIEDVAETSNLD-------QSMGGCDLDDSNDKTSADYYFDSY 69
++N+NNNNH D+ E +N D Q G + DD DK D D
Sbjct: 183 DSNDNNNNHPAVSATDDCSSQPKEAANDDNEPDNDVQFEGEEEEDDEEDKEEEDEEEDDE 362
Query: 70 SHFGNFHKEM 79
N E+
Sbjct: 363 DDDSNGEAEV 392
>TC8031 similar to UP|Q9FR33 (Q9FR33) Ripening regulated protein DDTFR10/A
(Fragment), partial (44%)
Length = 1109
Score = 23.1 bits (48), Expect(2) = 2.9
Identities = 8/14 (57%), Positives = 12/14 (85%)
Frame = +3
Query: 17 NNNNNNNNHLRFED 30
NN++NNNN+L F +
Sbjct: 195 NNHHNNNNNLGFNN 236
Score = 22.7 bits (47), Expect(2) = 2.9
Identities = 16/62 (25%), Positives = 25/62 (39%), Gaps = 1/62 (1%)
Frame = +2
Query: 28 FEDADEVIEDVAETSNLDQSMGGC-DLDDSNDKTSADYYFDSYSHFGNFHKEMLKDTVRT 86
F+ DE+ TSN + D S+ S D+YFD+ + F E +
Sbjct: 227 FQQFDEIT-----TSNTSPTQSSSSDSSSSSSNISLDHYFDTQNQLPLFEFEFKPHPIDL 391
Query: 87 KT 88
+T
Sbjct: 392 QT 397
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,196,201
Number of Sequences: 28460
Number of extensions: 63833
Number of successful extensions: 838
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 480
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 649
length of query: 322
length of database: 4,897,600
effective HSP length: 90
effective length of query: 232
effective length of database: 2,336,200
effective search space: 541998400
effective search space used: 541998400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC148360.2


