

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148347.6 + phase: 0
(807 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
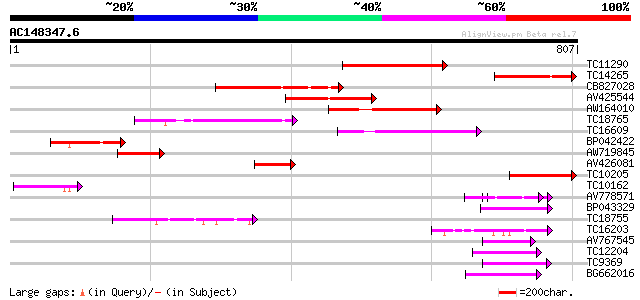
Score E
Sequences producing significant alignments: (bits) Value
TC11290 weakly similar to UP|AAN62760 (AAN62760) Disease resista... 189 1e-48
TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resista... 169 1e-42
CB827028 144 4e-35
AV425544 142 3e-34
AW164010 131 5e-31
TC18765 weakly similar to UP|Q9LVT1 (Q9LVT1) Disease resistance ... 125 3e-29
TC16609 weakly similar to PIR|T48468|T48468 disease resistance-l... 108 4e-24
BP042422 106 1e-23
AW719845 104 6e-23
AV426081 100 9e-22
TC10205 weakly similar to UP|Q8RL77 (Q8RL77) HpaG, partial (10%) 86 2e-17
TC10162 78 6e-15
AV778571 64 9e-11
BP043329 52 5e-07
TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fr... 52 5e-07
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 47 9e-06
AV767545 47 1e-05
TC12204 weakly similar to UP|Q9ZSN3 (Q9ZSN3) NBS-LRR-like protei... 45 5e-05
TC9369 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resista... 45 6e-05
BG662016 44 1e-04
>TC11290 weakly similar to UP|AAN62760 (AAN62760) Disease resistance
protein-like protein MsR1, partial (18%)
Length = 459
Score = 189 bits (481), Expect = 1e-48
Identities = 95/152 (62%), Positives = 119/152 (77%), Gaps = 2/152 (1%)
Frame = +3
Query: 474 GIYQSTKQPFEQRKRLIIDINKNRSE--LAEKQQSLLTRILSKVMRLCIKQNPQQLAAHI 531
GIYQS ++P +RKRL ID N+++ E L EKQQ +++R LSK+ RLC+KQ PQ + A
Sbjct: 3 GIYQSIQEPVGKRKRLNIDTNEDKLEWLLQEKQQGIMSRTLSKIRRLCLKQKPQLVLART 182
Query: 532 LSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCEL 591
LS+STDE C D S +QP + EVLILNL TK+YS PE +++M+KLK LI+TNY FHP EL
Sbjct: 183 LSISTDETCTPDLSLIQPAEAEVLILNLRTKKYSLPEILEEMNKLKALIVTNYGFHPSEL 362
Query: 592 NNFELLGSLQNLEKIRLERILVPSFGTLKNLK 623
NNFELL SL NL++IRLE+I VPSFGTLKNLK
Sbjct: 363 NNFELLDSLSNLKRIRLEQISVPSFGTLKNLK 458
>TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resistance
protein-like protein MsR1, partial (5%)
Length = 766
Score = 169 bits (429), Expect = 1e-42
Identities = 83/117 (70%), Positives = 98/117 (82%)
Frame = +1
Query: 690 QDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNL 749
Q+IGKLENLELL SSCTDL+ +P SIG+L L+ LDISNCISL SLPEE GNLCNLK+L
Sbjct: 1 QEIGKLENLELLRLSSCTDLKGLPDSIGRLSKLRLLDISNCISLPSLPEEIGNLCNLKSL 180
Query: 750 DMASCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLHVDVNLNWL 806
M SCA ELP S+VNLQNL T+ CDEETAA+WE F++++P++KIEV VDVNLNWL
Sbjct: 181 YMTSCAGCELPSSIVNLQNL-TVVCDEETAASWEAFEYVIPHLKIEVPQVDVNLNWL 348
Score = 32.0 bits (71), Expect = 0.40
Identities = 20/55 (36%), Positives = 27/55 (48%), Gaps = 1/55 (1%)
Frame = -3
Query: 709 LEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGN-LCNLKNLDMASCASIELPFS 762
L +P S GKL L D+S +S SLP E GN +++ L + S P S
Sbjct: 167 LHRLPISSGKLGRLMQXDMSRSLSFESLPIESGNPFKSVQELSLRSSKFSNFPIS 3
>CB827028
Length = 572
Score = 144 bits (364), Expect = 4e-35
Identities = 83/183 (45%), Positives = 120/183 (65%)
Frame = +2
Query: 293 ILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDD 352
+L+ L+ + A +LF H A + NS++I +LV+K+V+ CK P+ + V TSL+N+
Sbjct: 44 VLERLSLDYATSLFLHSASLSPNSTEI-PVSLVKKIVKGCKRSPIALTVTGTSLKNKNLA 220
Query: 353 LWRKIVMELSQGHSILDSNTELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAAL 412
+WR LS+G SIL+SN ++LT LQK DVL+ P +ECF D+ LFPED RIP AAL
Sbjct: 221 VWRSRAKALSKGQSILESNHDVLTCLQKSLDVLD--PIAIECFKDLGLFPEDQRIPAAAL 394
Query: 413 VDMWAKLYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRD 472
VDMWA+L DD AME I +L +N+A++ + RK A+D +YN H++ H +LR+
Sbjct: 395 VDMWAELRCGDDE--TAMEKIYELVNLNMADIFVTRKVANDI--VDYNYHYVAQHGLLRE 562
Query: 473 LGI 475
L I
Sbjct: 563 LAI 571
>AV425544
Length = 425
Score = 142 bits (357), Expect = 3e-34
Identities = 74/133 (55%), Positives = 95/133 (70%), Gaps = 3/133 (2%)
Frame = +1
Query: 393 ECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNLANVIIPRKNAS 452
ECFMD+ LFPED RIPV AL+DMWA+LY LD++G AM I+ L NL N I+ RK AS
Sbjct: 19 ECFMDLGLFPEDQRIPVTALIDMWAELYNLDEDGRNAMTIVLDLTSRNLINFIVTRKVAS 198
Query: 453 DTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDIN-KNRSE--LAEKQQSLLT 509
D YNNHF++LHD+LR+L I+QS +PFEQRKRLIID+N NR E + + QQ +
Sbjct: 199 DA-GVCYNNHFVMLHDLLRELAIHQSKGEPFEQRKRLIIDLNGDNRPEWWVGQNQQGFIG 375
Query: 510 RILSKVMRLCIKQ 522
R+ S + R+ +KQ
Sbjct: 376 RLFSFLPRMLVKQ 414
>AW164010
Length = 494
Score = 131 bits (329), Expect = 5e-31
Identities = 68/161 (42%), Positives = 103/161 (63%)
Frame = +2
Query: 454 TDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILS 513
+D +YN H++ H +LRDL I Q++++ ++R RLII+IN+N
Sbjct: 26 SDIVDYNYHYVTQHGLLRDLAILQTSEETEDKRDRLIIEINRNN---------------- 157
Query: 514 KVMRLCIKQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKM 573
+ +N +AA ILS+STDEA +S+ +QPT+VE L+LNL K+Y+ P ++KM
Sbjct: 158 -LPAWWSLENEYPIAARILSISTDEAFSSELCNLQPTKVEALVLNLRGKKYTLPMFMEKM 334
Query: 574 SKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERILVP 614
+KLKVL ITNY +P EL NFE+L L NL+++RLE++ +P
Sbjct: 335 NKLKVLTITNYDLYPAELENFEMLDHLSNLKRLRLEKVRIP 457
>TC18765 weakly similar to UP|Q9LVT1 (Q9LVT1) Disease resistance
protein-like, partial (18%)
Length = 688
Score = 125 bits (313), Expect = 3e-29
Identities = 83/242 (34%), Positives = 133/242 (54%), Gaps = 10/242 (4%)
Frame = +2
Query: 178 KTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEH--------CGYPVPEFQ 229
KTTLA ++C D +V FKE I+F+T S++P ++ + +I H Y VP++
Sbjct: 2 KTTLAREVCRDDQVRCHFKERILFLTVSQSPNVEELRAKIFGHIMGNRGLNANYAVPQWM 181
Query: 230 SDEDAVNKLELLLKKVEGSPLLLVLDDVWPTSESLVKKLQFEISDFKILVTSRVSFPR-F 288
+ ++ S +L+VLDDVW S ++++L + K LV SR F R F
Sbjct: 182 PQFECQSQ----------SQILVVLDDVW--SLPVLEQLVLRVPGCKYLVVSRFKFQRIF 325
Query: 289 RTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRN 348
T ++ L+ DA++LF H+A K+ ++NL+++VV C LPL +KVI SLR+
Sbjct: 326 NDTYDVELLSEGDALSLFCHHAFGHKSIPFGANQNLIKQVVAECGRLPLALKVIGASLRD 505
Query: 349 RPDDLWRKIVMELSQGHSILDS-NTELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRI 407
+ + W + LSQG SI +S L+ R+ + L + + ECF+D+ FPED +I
Sbjct: 506 QNEMFWLSVKTRLSQGLSIGESYEVNLIDRMAISTNYLPEK--VKECFLDLCAFPEDKKI 679
Query: 408 PV 409
P+
Sbjct: 680 PL 685
>TC16609 weakly similar to PIR|T48468|T48468 disease resistance-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(12%)
Length = 621
Score = 108 bits (269), Expect = 4e-24
Identities = 63/207 (30%), Positives = 101/207 (48%), Gaps = 2/207 (0%)
Frame = +1
Query: 467 HDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQ 526
HDILRDL + S + +R RL++ + +L ++ ++ Q
Sbjct: 43 HDILRDLALNLSNRGSINERLRLVMPKREGNGQLPKEW---------------LRYRGQP 177
Query: 527 LAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSF 586
L A I+S+ T E DW +++ + EVLILN + +Y P I +M L+ LI+ NYS
Sbjct: 178 LEARIVSIHTGEMTEGDWCELEFPKAEVLILNFTSSEYFLPPFIARMPSLRALIVINYST 357
Query: 587 HPCELNNFELLGSLQNLEKIRLERILVPSFG--TLKNLKKLSLYMCNTILAFEKGSILIS 644
L+N + +L NL + LE++ P L+ L KL + +C + + ++
Sbjct: 358 TYAHLHNVSVFKNLSNLRSLWLEKVSTPQLSGIVLEKLGKLFIVLCQVNNSLKGKDANLA 537
Query: 645 DAFPNLEELNIDYCKDLVVLQTGICDI 671
FPNL E +D+C DL L + IC I
Sbjct: 538 HIFPNLSEFTLDHCDDLTELPSSICGI 618
>BP042422
Length = 526
Score = 106 bits (265), Expect = 1e-23
Identities = 60/125 (48%), Positives = 80/125 (64%), Gaps = 19/125 (15%)
Frame = +3
Query: 59 HLDHPREEIKTLLEENDAEES-ACKC-----------------SSENDSYVVD-ENQSLI 99
HLD PREEIKTL+ E DA E C C ++++D VD E Q+ +
Sbjct: 156 HLDPPREEIKTLIREKDAGEGDKCLCWGNFISLLQLCVNKLCHNNKDDFLNVDGEKQAQM 335
Query: 100 VNDVEETLYKAREILELLNYETFEHKFNEAKPPFKRPFDVPENPKFTVGLDIPFSKLKME 159
NDV++TLYK R LEL++ E FE KF+ A P KRP+ VP NP+FTVG+D+P +KLK+E
Sbjct: 336 ANDVKDTLYKVRGFLELISKEDFEQKFSGA--PIKRPYGVPANPEFTVGVDVPLNKLKVE 509
Query: 160 LIRDG 164
L++DG
Sbjct: 510 LLKDG 524
>AW719845
Length = 209
Score = 104 bits (259), Expect = 6e-23
Identities = 46/67 (68%), Positives = 58/67 (85%)
Frame = +3
Query: 154 SKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLKTI 213
SKLKMEL++DG S + +TGLGGLGKTTLA +CWD+++ GKF+ENI+FVTFSKTP +K I
Sbjct: 6 SKLKMELLKDGVSVVAVTGLGGLGKTTLAAMVCWDKQIKGKFRENILFVTFSKTPNVKNI 185
Query: 214 VERIIEH 220
VER+ EH
Sbjct: 186 VERLFEH 206
>AV426081
Length = 179
Score = 100 bits (249), Expect = 9e-22
Identities = 47/59 (79%), Positives = 52/59 (87%)
Frame = +1
Query: 349 RPDDLWRKIVMELSQGHSILDSNTELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRI 407
RP +LW+KIV ELSQGHS+LDS+TELLTRLQKI DV EDNP I ECFMD+ALFPED RI
Sbjct: 1 RPYELWQKIVKELSQGHSVLDSHTELLTRLQKILDVAEDNPVIKECFMDLALFPEDQRI 177
>TC10205 weakly similar to UP|Q8RL77 (Q8RL77) HpaG, partial (10%)
Length = 586
Score = 85.9 bits (211), Expect = 2e-17
Identities = 42/96 (43%), Positives = 65/96 (66%), Gaps = 1/96 (1%)
Frame = +3
Query: 712 IPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASI-ELPFSVVNLQNLK 770
+P SI L ++ LDIS+CISLS LPE+ G L L+ L++ C+ + ELP S+++L+NL+
Sbjct: 9 LPKSITNLEQVQFLDISDCISLSHLPEDMGELKKLEKLNVRGCSRLTELPTSIMDLENLE 188
Query: 771 TITCDEETAATWEDFQHMLPNMKIEVLHVDVNLNWL 806
+ CDEE A WE Q +L ++++ V D NL++L
Sbjct: 189 EVECDEEIAELWEPVQQILSDLQLHVAPTDFNLDFL 296
Score = 37.7 bits (86), Expect = 0.007
Identities = 23/54 (42%), Positives = 31/54 (56%), Gaps = 3/54 (5%)
Frame = +3
Query: 649 NLEE---LNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLE 699
NLE+ L+I C L L + ++ L+KLNV C +L+ LP I LENLE
Sbjct: 27 NLEQVQFLDISDCISLSHLPEDMGELKKLEKLNVRGCSRLTELPTSIMDLENLE 188
>TC10162
Length = 577
Score = 77.8 bits (190), Expect = 6e-15
Identities = 49/118 (41%), Positives = 65/118 (54%), Gaps = 20/118 (16%)
Frame = +1
Query: 6 THIITLT--TRFHDVLNKISQIVETARKHQSARQLLRSILKDLTLVVQDIKQYNEHLDHP 63
T ++ L RF ++L KISQ+VE +RK A+++LRS L + VV +IK+YNEHL P
Sbjct: 211 TQLVALAPLARFQEILKKISQMVEKSRKSGIAQRILRSTLIEFNHVVLEIKRYNEHLAQP 390
Query: 64 REEIKTLLEENDA---EESACKC---------------SSENDSYVVDENQSLIVNDV 103
REEIKTL+ E DA E KC ++DS DE Q+L DV
Sbjct: 391 REEIKTLIREKDAGEEGEEGDKCVNSCLQVVTDRVGHGKDDSDSVDADEKQALTAKDV 564
>AV778571
Length = 452
Score = 63.9 bits (154), Expect = 9e-11
Identities = 34/113 (30%), Positives = 62/113 (54%)
Frame = +3
Query: 648 PNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCT 707
PNLE+L+ Y ++L+ + + + L+ L++ C K+ + P KL +LE L S C+
Sbjct: 84 PNLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPM--KLASLEELYLSHCS 257
Query: 708 DLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIELP 760
LE+ P + K+ N+ + + + + LP GNL L+ L++ C ++LP
Sbjct: 258 SLESFPEILAKMENITLIGVYHT-PIKELPSSIGNLTRLRRLELHGCGMLQLP 413
Score = 48.9 bits (115), Expect = 3e-06
Identities = 29/100 (29%), Positives = 48/100 (48%)
Frame = +3
Query: 673 SLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCIS 732
+L+KL+ L + + +G L L +L C + P KL +L+ L +S+C S
Sbjct: 87 NLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPM--KLASLEELYLSHCSS 260
Query: 733 LSSLPEEFGNLCNLKNLDMASCASIELPFSVVNLQNLKTI 772
L S PE + N+ + + ELP S+ NL L+ +
Sbjct: 261 LESFPEILAKMENITLIGVYHTPIKELPSSIGNLTRLRRL 380
Score = 47.0 bits (110), Expect = 1e-05
Identities = 30/94 (31%), Positives = 50/94 (52%), Gaps = 1/94 (1%)
Frame = +3
Query: 680 TNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEE 739
+ C ++ +P D+ L NLE LSF +L I S+G L L+ LDI C + + P
Sbjct: 39 SECESITQIP-DLSGLPNLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPM 215
Query: 740 FGNLCNLKNLDMASCASIE-LPFSVVNLQNLKTI 772
L +L+ L ++ C+S+E P + ++N+ I
Sbjct: 216 --KLASLEELYLSHCSSLESFPEILAKMENITLI 311
>BP043329
Length = 484
Score = 51.6 bits (122), Expect = 5e-07
Identities = 34/104 (32%), Positives = 56/104 (53%), Gaps = 2/104 (1%)
Frame = +3
Query: 671 IISLKKLNVT-NCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISN 729
+ +LK LN++ N +P +IG L NLE++ + C + IP SIG L LK LD++
Sbjct: 27 LTTLKMLNLSYNPFYPGRIPPEIGNLTNLEVVWLTQCNLVGVIPDSIGNLKKLKDLDLAL 206
Query: 730 CISLSSLPEEFGNLCNLKNLDM-ASCASIELPFSVVNLQNLKTI 772
S+P L +L+ +++ + S ELP + NL L+ +
Sbjct: 207 NDLYGSIPSSLTGLTSLRQIELYNNSLSGELPRGMGNLTELRLL 338
Score = 40.0 bits (92), Expect = 0.001
Identities = 44/158 (27%), Positives = 72/158 (44%), Gaps = 2/158 (1%)
Frame = +3
Query: 614 PSFGTLKNLKKLSLYMCNTILAFEKGSILISDA-FPNLEELNIDYCKDLVVLQTGICDII 672
PS GTL LK L+L + F G I NLE + + C + V+ I ++
Sbjct: 12 PSLGTLTTLKMLNL----SYNPFYPGRIPPEIGNLTNLEVVWLTQCNLVGVIPDSIGNLK 179
Query: 673 SLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCIS 732
LK L++ S+P + L +L + + + +P +G L L+ LD S
Sbjct: 180 KLKDLDLALNDLYGSIPSSLTGLTSLRQIELYNNSLSGELPRGMGNLTELRLLDASMNHL 359
Query: 733 LSSLPEEFGNLCNLKNLDM-ASCASIELPFSVVNLQNL 769
+PEE +L L++L++ + ELP S+ + NL
Sbjct: 360 TGRIPEELCSL-PLESLNLYENRFEGELPASIADSPNL 470
>TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fragment),
complete
Length = 880
Score = 51.6 bits (122), Expect = 5e-07
Identities = 62/229 (27%), Positives = 110/229 (47%), Gaps = 23/229 (10%)
Frame = +3
Query: 147 VGLDIPFSKLKMELIRDGSSTLVL--TGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTF 204
VG+D KL LI+ V+ TG+GG+GKTTL K +D V K ++T
Sbjct: 198 VGIDRRKKKLMGCLIKPCPVRKVISVTGMGGMGKTTL-VKQVYDDPVVIKHFRACAWITV 374
Query: 205 SKT----PMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLKK-VEGSPLLLVLDDVWP 259
S++ +L+ + ++ PVP + ++L++++K ++ L+V DDVW
Sbjct: 375 SQSCEIGELLRDLARQLFSEIRRPVP-LGLENMRCDRLKMIIKDLLQRRRYLVVFDDVWH 551
Query: 260 TSESLVKKLQFEISD----FKILVTSRVSFPRFRTTC-------ILKPLAHEDAVTLF-H 307
E + +++ + D +I++T+R S F ++ L+PL ++A LF
Sbjct: 552 VRE--WEAVKYALPDNNCGSRIMITTRRSDLAFTSSTESKGKVYNLQPLKEDEAWELFCR 725
Query: 308 HYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTI----KVIATSLRNRPDD 352
+ S +I + ++R C+GLPL I V+AT + R D+
Sbjct: 726 KTFHGDSCPSHLI--GICTYILRKCEGLPLAIVAISGVLATKDKRRIDE 866
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation
aberrant root formation protein), complete
Length = 3308
Score = 47.4 bits (111), Expect = 9e-06
Identities = 63/209 (30%), Positives = 95/209 (45%), Gaps = 37/209 (17%)
Frame = +2
Query: 601 QNLEKIRLERILVPSFG-------TLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEEL 653
QNL + L LVP FG L+ L+ L++ M N L + S L S +L+ L
Sbjct: 323 QNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNN--LTDQLPSDLAS--LTSLKVL 490
Query: 654 NIDYCKDLVVLQTGICDIISLKKLNVTNCHKLS---SLPQDIGKLENLELL--------- 701
NI + +L Q + + +L + + S LP++I KLE L+ L
Sbjct: 491 NISH--NLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSG 664
Query: 702 ----SFSSCTDLE-----------AIPTSIGKLLNLK--HLDISNCISLSSLPEEFGNLC 744
S+S LE +P S+ KL LK HL SN +P FG++
Sbjct: 665 TIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYE-GGIPPAFGSME 841
Query: 745 NLKNLDMASC-ASIELPFSVVNLQNLKTI 772
NL+ L+MA+C + E+P S+ NL L ++
Sbjct: 842 NLRLLEMANCNLTGEIPPSLGNLTKLHSL 928
Score = 33.1 bits (74), Expect = 0.18
Identities = 62/277 (22%), Positives = 104/277 (37%), Gaps = 59/277 (21%)
Frame = +2
Query: 511 ILSKVMRLCIKQN------PQQLAA----HILSVSTDEACASDWSQMQPTQVEVLILNLH 560
+L K+ L I N P LA+ +L++S + + E+ L+ +
Sbjct: 395 LLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMTELEALDAY 574
Query: 561 TKQYS--FPESIKKMSKLKVLIITNYSFHPC------ELNNFELLG-------------- 598
+S PE I K+ KLK L + F E + E LG
Sbjct: 575 DNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESL 754
Query: 599 -SLQNLEKIRL------ERILVPSFGTLKNLKKLSLYMCNTI--LAFEKGSIL-ISDAFP 648
L+ L+++ L E + P+FG+++NL+ L + CN + G++ + F
Sbjct: 755 AKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFV 934
Query: 649 NLEELN------IDYCKDLVVLQTGICDIIS--------LKKLNVTNCHK---LSSLPQD 691
+ L + L+ L I D+ LK L + N + SLP
Sbjct: 935 QMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSF 1114
Query: 692 IGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDIS 728
IG L NLE L +P ++G + D++
Sbjct: 1115IGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVT 1225
>AV767545
Length = 562
Score = 47.0 bits (110), Expect = 1e-05
Identities = 29/76 (38%), Positives = 42/76 (55%)
Frame = -1
Query: 673 SLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCIS 732
SLK LN+ + + +P I L +LE L S L AIP+S+G+LL L+ LD+S
Sbjct: 538 SLKVLNLGSNNISGPIPVSISNLIDLERLDISRNHILGAIPSSLGQLLELQWLDVSINSL 359
Query: 733 LSSLPEEFGNLCNLKN 748
S+P + NLK+
Sbjct: 358 AGSIPSSLSQITNLKH 311
>TC12204 weakly similar to UP|Q9ZSN3 (Q9ZSN3) NBS-LRR-like protein cD7
(Fragment), partial (3%)
Length = 328
Score = 45.1 bits (105), Expect = 5e-05
Identities = 31/100 (31%), Positives = 48/100 (48%), Gaps = 1/100 (1%)
Frame = +1
Query: 659 KDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGK 718
+ LVV + + SL L + NC S P+ + N+ L C L++ P + K
Sbjct: 10 ESLVVTGVQLQYLQSLNSLRICNCPNFESFPEGGLRAPNMTNLHLEKCKKLKSFPQQMNK 189
Query: 719 -LLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASI 757
LL+L L+I C L S+PE G +L L++ CA +
Sbjct: 190 MLLSLMTLNIKECPELESIPEG-GFPDSLNLLEIFHCAKL 306
>TC9369 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resistance
protein-like protein, partial (70%)
Length = 859
Score = 44.7 bits (104), Expect = 6e-05
Identities = 31/98 (31%), Positives = 52/98 (52%), Gaps = 1/98 (1%)
Frame = +1
Query: 674 LKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISL 733
L +L++ N +P IG+L+ L++L+ +AIP IG+L +L HL +S
Sbjct: 13 LTRLDLHNNKLTGPIPPQIGRLKRLKILNLRWNKLQDAIPPEIGELKSLTHLYLSFNSFK 192
Query: 734 SSLPEEFGNLCNLKNLDMASCASI-ELPFSVVNLQNLK 770
+P+E NL +L+ L + I +P + LQNL+
Sbjct: 193 GEIPKELANLPDLRYLYLHENRLIGRIPPELGTLQNLR 306
>BG662016
Length = 459
Score = 43.9 bits (102), Expect = 1e-04
Identities = 38/108 (35%), Positives = 58/108 (53%), Gaps = 1/108 (0%)
Frame = +1
Query: 650 LEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDL 709
LEELN ++ K + T ++I+LKKL V N +KL LP L L +L + L
Sbjct: 133 LEELNANFNKLSKLPDTIGFELINLKKLAV-NSNKLVLLPSSTSHLTALTVLD-ARLNCL 306
Query: 710 EAIPTSIGKLLNLKHLDIS-NCISLSSLPEEFGNLCNLKNLDMASCAS 756
A+P ++ L+NL+ L++S N L +LP G L + N +A+ S
Sbjct: 307 RALPENLENLINLETLNVSQNFRYLDTLPYSIGLLLSWLNWMLATTIS 450
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,717,422
Number of Sequences: 28460
Number of extensions: 184732
Number of successful extensions: 944
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 876
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 912
length of query: 807
length of database: 4,897,600
effective HSP length: 98
effective length of query: 709
effective length of database: 2,108,520
effective search space: 1494940680
effective search space used: 1494940680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148347.6


