

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148347.4 + phase: 1 /pseudo
(776 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
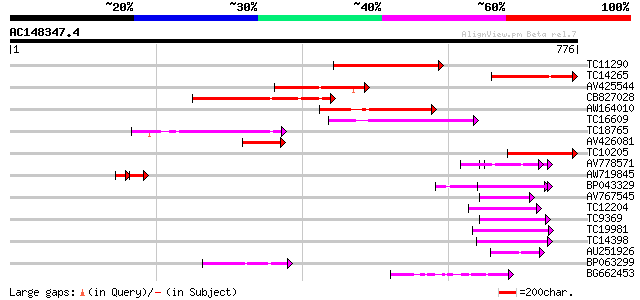
Score E
Sequences producing significant alignments: (bits) Value
TC11290 weakly similar to UP|AAN62760 (AAN62760) Disease resista... 196 2e-50
TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resista... 166 1e-41
AV425544 149 2e-36
CB827028 148 3e-36
AW164010 123 1e-28
TC16609 weakly similar to PIR|T48468|T48468 disease resistance-l... 112 2e-25
TC18765 weakly similar to UP|Q9LVT1 (Q9LVT1) Disease resistance ... 111 4e-25
AV426081 104 6e-23
TC10205 weakly similar to UP|Q8RL77 (Q8RL77) HpaG, partial (10%) 85 4e-17
AV778571 65 5e-11
AW719845 42 1e-07
BP043329 53 2e-07
AV767545 48 7e-06
TC12204 weakly similar to UP|Q9ZSN3 (Q9ZSN3) NBS-LRR-like protei... 46 2e-05
TC9369 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resista... 46 3e-05
TC19981 weakly similar to UP|AAP20228 (AAP20228) Resistance prot... 44 8e-05
TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%) 44 8e-05
AU251926 44 1e-04
BP063299 44 1e-04
BG662453 44 1e-04
>TC11290 weakly similar to UP|AAN62760 (AAN62760) Disease resistance
protein-like protein MsR1, partial (18%)
Length = 459
Score = 196 bits (497), Expect = 2e-50
Identities = 101/152 (66%), Positives = 121/152 (79%), Gaps = 2/152 (1%)
Frame = +3
Query: 444 GIYQSTKEPFEQRKRLIIDMNNNKSG--LAEKQQGLMTRIFSKFMRLCVKQNPQQLAARI 501
GIYQS +EP +RKRL ID N +K L EKQQG+M+R SK RLC+KQ PQ + AR
Sbjct: 3 GIYQSIQEPVGKRKRLNIDTNEDKLEWLLQEKQQGIMSRTLSKIRRLCLKQKPQLVLART 182
Query: 502 LSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKL 561
LS+STDETC D S +QPA+ EVLILN+ TK+YSLPE + +M+KL+ LI+TNYGFHPS+L
Sbjct: 183 LSISTDETCTPDLSLIQPAEAEVLILNLRTKKYSLPEILEEMNKLKALIVTNYGFHPSEL 362
Query: 562 NNIELLGSLQNLERIRLERISVPSFGTLKNLK 593
NN ELL SL NL+RIRLE+ISVPSFGTLKNLK
Sbjct: 363 NNFELLDSLSNLKRIRLEQISVPSFGTLKNLK 458
>TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resistance
protein-like protein MsR1, partial (5%)
Length = 766
Score = 166 bits (421), Expect = 1e-41
Identities = 82/117 (70%), Positives = 98/117 (83%)
Frame = +1
Query: 660 QDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNL 719
Q+IGKLENLELL LSSCTDL+ +P SIG+L L+ LDISNCISL SLPEE GNLCNLK+L
Sbjct: 1 QEIGKLENLELLRLSSCTDLKGLPDSIGRLSKLRLLDISNCISLPSLPEEIGNLCNLKSL 180
Query: 720 DMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLHNMKIEVLHVDVNLNWL 776
M +CA ELP S+VNLQNL T+ CDEETAA+WE F++++ ++KIEV VDVNLNWL
Sbjct: 181 YMTSCAGCELPSSIVNLQNL-TVVCDEETAASWEAFEYVIPHLKIEVPQVDVNLNWL 348
Score = 31.6 bits (70), Expect = 0.50
Identities = 25/76 (32%), Positives = 42/76 (54%)
Frame = +1
Query: 522 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERI 581
+E+L L+ T LP+ I ++SKLR+L I+N PS I L +L++L
Sbjct: 25 LELLRLSSCTDLKGLPDSIGRLSKLRLLDISNCISLPSLPEEIGNLCNLKSLYMTSCAGC 204
Query: 582 SVPSFGTLKNLKKLSL 597
+PS ++ NL+ L++
Sbjct: 205 ELPS--SIVNLQNLTV 246
Score = 31.2 bits (69), Expect = 0.66
Identities = 16/34 (47%), Positives = 19/34 (55%)
Frame = -3
Query: 679 LEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGN 712
L +P S GKL L D+S +S SLP E GN
Sbjct: 167 LHRLPISSGKLGRLMQXDMSRSLSFESLPIESGN 66
>AV425544
Length = 425
Score = 149 bits (375), Expect = 2e-36
Identities = 78/133 (58%), Positives = 94/133 (70%), Gaps = 3/133 (2%)
Frame = +1
Query: 363 ECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNAS 422
ECFMDL LFPED RIPV AL+DMWAELY LD++G AM I+ L NL N I+ RK AS
Sbjct: 19 ECFMDLGLFPEDQRIPVTALIDMWAELYNLDEDGRNAMTIVLDLTSRNLINFIVTRKVAS 198
Query: 423 DTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKS---GLAEKQQGLMT 479
D YNNHF++LHD+LREL I+QS EPFEQRKRLIID+N + + + QQG +
Sbjct: 199 DA-GVCYNNHFVMLHDLLRELAIHQSKGEPFEQRKRLIIDLNGDNRPEWWVGQNQQGFIG 375
Query: 480 RIFSKFMRLCVKQ 492
R+FS R+ VKQ
Sbjct: 376 RLFSFLPRMLVKQ 414
>CB827028
Length = 572
Score = 148 bits (374), Expect = 3e-36
Identities = 90/197 (45%), Positives = 125/197 (62%), Gaps = 2/197 (1%)
Frame = +2
Query: 251 SRVAFPRFSTTC--ILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLT 308
SR A R+ + +L+ L+ + A +LF H A + NS++I +LV+K+V+ C P+
Sbjct: 2 SRSAIARYDSGSPYVLERLSLDYATSLFLHSASLSPNSTEI-PVSLVKKIVKGCKRSPIA 178
Query: 309 IKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMDL 368
+ V TSLKN+ +W K LS+G SIL+SN ++LT LQK DVL+ P +ECF DL
Sbjct: 179 LTVTGTSLKNKNLAVWRSRAKALSKGQSILESNHDVLTCLQKSLDVLD--PIAIECFKDL 352
Query: 369 ALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNN 428
LFPED RIP AALVDMWAEL DD AME I +L +N+A++ + RK A+D +
Sbjct: 353 GLFPEDQRIPAAALVDMWAELRCGDDE--TAMEKIYELVNLNMADIFVTRKVANDI--VD 520
Query: 429 YNNHFIILHDILRELGI 445
YN H++ H +LREL I
Sbjct: 521 YNYHYVAQHGLLRELAI 571
>AW164010
Length = 494
Score = 123 bits (308), Expect = 1e-28
Identities = 64/161 (39%), Positives = 103/161 (63%)
Frame = +2
Query: 424 TDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFS 483
+D +YN H++ H +LR+L I Q+++E ++R RLII++N N
Sbjct: 26 SDIVDYNYHYVTQHGLLRDLAILQTSEETEDKRDRLIIEINRNN--------------LP 163
Query: 484 KFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKM 543
+ L +N +AARILS+STDE + + ++QP +VE L+LN+ K+Y+LP ++ KM
Sbjct: 164 AWWSL---ENEYPIAARILSISTDEAFSSELCNLQPTKVEALVLNLRGKKYTLPMFMEKM 334
Query: 544 SKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVP 584
+KL+VL ITNY +P++L N E+L L NL+R+RLE++ +P
Sbjct: 335 NKLKVLTITNYDLYPAELENFEMLDHLSNLKRLRLEKVRIP 457
>TC16609 weakly similar to PIR|T48468|T48468 disease resistance-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(12%)
Length = 621
Score = 112 bits (280), Expect = 2e-25
Identities = 66/207 (31%), Positives = 104/207 (49%), Gaps = 2/207 (0%)
Frame = +1
Query: 437 HDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQ 496
HDILR+L + S + +R RL++ L ++ ++ Q
Sbjct: 43 HDILRDLALNLSNRGSINERLRLVMPKREGNGQLPKEW---------------LRYRGQP 177
Query: 497 LAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGF 556
L ARI+S+ T E DW ++ + EVLILN + +Y LP +IA+M LR LI+ NY
Sbjct: 178 LEARIVSIHTGEMTEGDWCELEFPKAEVLILNFTSSEYFLPPFIARMPSLRALIVINYST 357
Query: 557 HPSKLNNIELLGSLQNLERIRLERISVPSFG--TLKNLKKLSLYMCNTILAFEKGSILIS 614
+ L+N+ + +L NL + LE++S P L+ L KL + +C + + ++
Sbjct: 358 TYAHLHNVSVFKNLSNLRSLWLEKVSTPQLSGIVLEKLGKLFIVLCQVNNSLKGKDANLA 537
Query: 615 DAFPNLEELNIDYCKDLVVLQTGICDI 641
FPNL E +D+C DL L + IC I
Sbjct: 538 HIFPNLSEFTLDHCDDLTELPSSICGI 618
>TC18765 weakly similar to UP|Q9LVT1 (Q9LVT1) Disease resistance
protein-like, partial (18%)
Length = 688
Score = 111 bits (278), Expect = 4e-25
Identities = 76/223 (34%), Positives = 124/223 (55%), Gaps = 10/223 (4%)
Frame = +2
Query: 167 ENIIFVTFSKTPMLKTIVERIHQH--------CGYSVPEFQSDEDAVNKLGLLLKKVEGS 218
E I+F+T S++P ++ + +I H Y+VP++ + ++ S
Sbjct: 59 ERILFLTVSQSPNVEELRAKIFGHIMGNRGLNANYAVPQWMPQFECQSQ----------S 208
Query: 219 PLLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPR-FSTTCILKPLAHEDAVTLFH 277
+L+VLDDVW S ++E+L ++ G K LV SR F R F+ T ++ L+ DA++LF
Sbjct: 209 QILVVLDDVW--SLPVLEQLVLRVPGCKYLVVSRFKFQRIFNDTYDVELLSEGDALSLFC 382
Query: 278 HYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKNRPHDLWHKIVKELSQGHSI 337
H+A K+ ++NL+++VV C LPL +KVI SL+++ W + LSQG SI
Sbjct: 383 HHAFGHKSIPFGANQNLIKQVVAECGRLPLALKVIGASLRDQNEMFWLSVKTRLSQGLSI 562
Query: 338 LDS-NTELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPV 379
+S L+ R+ + L + K+ ECF+DL FPED +IP+
Sbjct: 563 GESYEVNLIDRMAISTNYLPE--KVKECFLDLCAFPEDKKIPL 685
>AV426081
Length = 179
Score = 104 bits (259), Expect = 6e-23
Identities = 49/59 (83%), Positives = 53/59 (89%)
Frame = +1
Query: 319 RPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRI 377
RP++LW KIVKELSQGHS+LDS+TELLTRLQKI DV EDNP I ECFMDLALFPED RI
Sbjct: 1 RPYELWQKIVKELSQGHSVLDSHTELLTRLQKILDVAEDNPVIKECFMDLALFPEDQRI 177
>TC10205 weakly similar to UP|Q8RL77 (Q8RL77) HpaG, partial (10%)
Length = 586
Score = 85.1 bits (209), Expect = 4e-17
Identities = 42/96 (43%), Positives = 65/96 (66%), Gaps = 1/96 (1%)
Frame = +3
Query: 682 IPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI-ELPFSVVNLQNLK 740
+P SI L ++ LDIS+CISLS LPE+ G L L+ L++ C+ + ELP S+++L+NL+
Sbjct: 9 LPKSITNLEQVQFLDISDCISLSHLPEDMGELKKLEKLNVRGCSRLTELPTSIMDLENLE 188
Query: 741 TITCDEETAATWEDFQHMLHNMKIEVLHVDVNLNWL 776
+ CDEE A WE Q +L ++++ V D NL++L
Sbjct: 189 EVECDEEIAELWEPVQQILSDLQLHVAPTDFNLDFL 296
Score = 37.7 bits (86), Expect = 0.007
Identities = 23/54 (42%), Positives = 31/54 (56%), Gaps = 3/54 (5%)
Frame = +3
Query: 619 NLEE---LNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLE 669
NLE+ L+I C L L + ++ L+KLNV C +L+ LP I LENLE
Sbjct: 27 NLEQVQFLDISDCISLSHLPEDMGELKKLEKLNVRGCSRLTELPTSIMDLENLE 188
Score = 27.3 bits (59), Expect = 9.5
Identities = 29/109 (26%), Positives = 44/109 (39%), Gaps = 12/109 (11%)
Frame = -1
Query: 616 AFPNLEELNIDY------------CKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIG 663
A PN +E N++ C+ L + TG + S + + K S D+G
Sbjct: 328 ANPNYDERNLERKSKLKSVGATCSCRSLRICWTGSHN--SAISSSHSTSSKFSRSMIDVG 155
Query: 664 KLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGN 712
NLE S ++ P S GK L L ++S + S +FGN
Sbjct: 154 NSVNLEQPLTLSFSNFFNSPMSSGK*LRLMQSEMSRNCTCSRFVIDFGN 8
>AV778571
Length = 452
Score = 64.7 bits (156), Expect = 5e-11
Identities = 35/113 (30%), Positives = 63/113 (54%)
Frame = +3
Query: 618 PNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCT 677
PNLE+L+ Y ++L+ + + + L+ L++ C K+ + P KL +LE L LS C+
Sbjct: 84 PNLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPM--KLASLEELYLSHCS 257
Query: 678 DLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIELP 730
LE+ P + K+ N+ + + + + LP GNL L+ L++ C ++LP
Sbjct: 258 SLESFPEILAKMENITLIGVYHT-PIKELPSSIGNLTRLRRLELHGCGMLQLP 413
Score = 49.3 bits (116), Expect = 2e-06
Identities = 29/100 (29%), Positives = 49/100 (49%)
Frame = +3
Query: 643 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCIS 702
+L+KL+ L + + +G L L +L + C + P KL +L+ L +S+C S
Sbjct: 87 NLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPM--KLASLEELYLSHCSS 260
Query: 703 LSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTI 742
L S PE + N+ + + ELP S+ NL L+ +
Sbjct: 261 LESFPEILAKMENITLIGVYHTPIKELPSSIGNLTRLRRL 380
Score = 44.3 bits (103), Expect = 8e-05
Identities = 29/94 (30%), Positives = 49/94 (51%), Gaps = 1/94 (1%)
Frame = +3
Query: 650 TNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEE 709
+ C ++ +P D+ L NLE LS +L I S+G L L+ LDI C + + P
Sbjct: 39 SECESITQIP-DLSGLPNLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPM 215
Query: 710 FGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTI 742
L +L+ L ++ C+S+E P + ++N+ I
Sbjct: 216 --KLASLEELYLSHCSSLESFPEILAKMENITLI 311
>AW719845
Length = 209
Score = 41.6 bits (96), Expect(2) = 1e-07
Identities = 18/27 (66%), Positives = 22/27 (80%)
Frame = +3
Query: 164 KLMENIIFVTFSKTPMLKTIVERIHQH 190
K ENI+FVTFSKTP +K IVER+ +H
Sbjct: 126 KFRENILFVTFSKTPNVKNIVERLFEH 206
Score = 31.6 bits (70), Expect(2) = 1e-07
Identities = 14/20 (70%), Positives = 15/20 (75%)
Frame = +1
Query: 146 V*EDWGKPLWLQSFVGMKKL 165
V* WGK LWLQ FVG+ KL
Sbjct: 61 V*VGWGKQLWLQWFVGINKL 120
>BP043329
Length = 484
Score = 52.8 bits (125), Expect = 2e-07
Identities = 35/104 (33%), Positives = 56/104 (53%), Gaps = 2/104 (1%)
Frame = +3
Query: 641 IISLKKLNVT-NCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISN 699
+ +LK LN++ N +P +IG L NLE++ L+ C + IP SIG L LK LD++
Sbjct: 27 LTTLKMLNLSYNPFYPGRIPPEIGNLTNLEVVWLTQCNLVGVIPDSIGNLKKLKDLDLAL 206
Query: 700 CISLSSLPEEFGNLCNLKNLDM-ATCASIELPFSVVNLQNLKTI 742
S+P L +L+ +++ S ELP + NL L+ +
Sbjct: 207 NDLYGSIPSSLTGLTSLRQIELYNNSLSGELPRGMGNLTELRLL 338
Score = 41.2 bits (95), Expect = 6e-04
Identities = 45/158 (28%), Positives = 72/158 (45%), Gaps = 2/158 (1%)
Frame = +3
Query: 584 PSFGTLKNLKKLSLYMCNTILAFEKGSILISDA-FPNLEELNIDYCKDLVVLQTGICDII 642
PS GTL LK L+L + F G I NLE + + C + V+ I ++
Sbjct: 12 PSLGTLTTLKMLNL----SYNPFYPGRIPPEIGNLTNLEVVWLTQCNLVGVIPDSIGNLK 179
Query: 643 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCIS 702
LK L++ S+P + L +L + L + + +P +G L L+ LD S
Sbjct: 180 KLKDLDLALNDLYGSIPSSLTGLTSLRQIELYNNSLSGELPRGMGNLTELRLLDASMNHL 359
Query: 703 LSSLPEEFGNLCNLKNLDM-ATCASIELPFSVVNLQNL 739
+PEE +L L++L++ ELP S+ + NL
Sbjct: 360 TGRIPEELCSL-PLESLNLYENRFEGELPASIADSPNL 470
>AV767545
Length = 562
Score = 47.8 bits (112), Expect = 7e-06
Identities = 29/76 (38%), Positives = 43/76 (56%)
Frame = -1
Query: 643 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCIS 702
SLK LN+ + + +P I L +LE L +S L AIP+S+G+LL L+ LD+S
Sbjct: 538 SLKVLNLGSNNISGPIPVSISNLIDLERLDISRNHILGAIPSSLGQLLELQWLDVSINSL 359
Query: 703 LSSLPEEFGNLCNLKN 718
S+P + NLK+
Sbjct: 358 AGSIPSSLSQITNLKH 311
>TC12204 weakly similar to UP|Q9ZSN3 (Q9ZSN3) NBS-LRR-like protein cD7
(Fragment), partial (3%)
Length = 328
Score = 46.2 bits (108), Expect = 2e-05
Identities = 32/100 (32%), Positives = 49/100 (49%), Gaps = 1/100 (1%)
Frame = +1
Query: 629 KDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGK 688
+ LVV + + SL L + NC S P+ + N+ L L C L++ P + K
Sbjct: 10 ESLVVTGVQLQYLQSLNSLRICNCPNFESFPEGGLRAPNMTNLHLEKCKKLKSFPQQMNK 189
Query: 689 -LLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI 727
LL+L L+I C L S+PE G +L L++ CA +
Sbjct: 190 MLLSLMTLNIKECPELESIPEG-GFPDSLNLLEIFHCAKL 306
Score = 40.0 bits (92), Expect = 0.001
Identities = 30/110 (27%), Positives = 54/110 (48%), Gaps = 1/110 (0%)
Frame = +1
Query: 571 QNLERIRLERISVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKD 630
QNLE + + + + L++L L + C +F +G + PN+ L+++ CK
Sbjct: 1 QNLESLVVTGVQLQY---LQSLNSLRICNCPNFESFPEGGLRA----PNMTNLHLEKCKK 159
Query: 631 LVVLQTGICD-IISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDL 679
L + ++SL LN+ C +L S+P+ G ++L LL + C L
Sbjct: 160 LKSFPQQMNKMLLSLMTLNIKECPELESIPEG-GFPDSLNLLEIFHCAKL 306
>TC9369 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resistance
protein-like protein, partial (70%)
Length = 859
Score = 45.8 bits (107), Expect = 3e-05
Identities = 32/98 (32%), Positives = 53/98 (53%), Gaps = 1/98 (1%)
Frame = +1
Query: 644 LKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISL 703
L +L++ N +P IG+L+ L++L+L +AIP IG+L +L HL +S
Sbjct: 13 LTRLDLHNNKLTGPIPPQIGRLKRLKILNLRWNKLQDAIPPEIGELKSLTHLYLSFNSFK 192
Query: 704 SSLPEEFGNLCNLKNLDMATCASI-ELPFSVVNLQNLK 740
+P+E NL +L+ L + I +P + LQNL+
Sbjct: 193 GEIPKELANLPDLRYLYLHENRLIGRIPPELGTLQNLR 306
>TC19981 weakly similar to UP|AAP20228 (AAP20228) Resistance protein SlVe1
precursor, partial (9%)
Length = 403
Score = 44.3 bits (103), Expect = 8e-05
Identities = 32/112 (28%), Positives = 55/112 (48%), Gaps = 1/112 (0%)
Frame = +1
Query: 634 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLK 693
+ + + D+ SL L +++ + S++P NL L L +C+ P I + L+
Sbjct: 67 IDSSLADLKSLSILQLSHNNLSSTVPDSFANFSNLTTLHLINCSLNGYFPKEIFHIKTLE 246
Query: 694 HLDISNCISLSSLPEEFGNLCNLKNLDMA-TCASIELPFSVVNLQNLKTITC 744
LD+S L F +L NLD++ T S ++P S+ NL+ L I+C
Sbjct: 247 VLDVSENKFLHGSFPNFTPSWSLHNLDVSHTNFSGQIPESISNLKKLTKISC 402
>TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%)
Length = 982
Score = 44.3 bits (103), Expect = 8e-05
Identities = 31/105 (29%), Positives = 52/105 (49%), Gaps = 1/105 (0%)
Frame = +1
Query: 639 CDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDIS 698
C S+ ++++ N + L D+G L +L+ L L IP +G L +L LD+
Sbjct: 313 CQDNSVTRVDLGNLNLSGHLVPDLGNLHSLQYLELYENNIQGTIPEELGNLQSLISLDLY 492
Query: 699 NCISLSSLPEEFGNLCNLKNLDM-ATCASIELPFSVVNLQNLKTI 742
+ S+P GNL NL+ L + + ++P S+ L NLK +
Sbjct: 493 HNNVSGSIPSSLGNLKNLRFLRLNNNHLTGQIPKSLSTLPNLKVL 627
Score = 40.0 bits (92), Expect = 0.001
Identities = 38/128 (29%), Positives = 55/128 (42%)
Frame = +1
Query: 572 NLERIRLERISVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDL 631
+L + L VP G L +L+ L LY N I P EEL
Sbjct: 340 DLGNLNLSGHLVPDLGNLHSLQYLELYENN-----------IQGTIP--EELG------- 459
Query: 632 VVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLN 691
++ SL L++ + + S+P +G L+NL L L++ IP S+ L N
Sbjct: 460 --------NLQSLISLDLYHNNVSGSIPSSLGNLKNLRFLRLNNNHLTGQIPKSLSTLPN 615
Query: 692 LKHLDISN 699
LK LD+SN
Sbjct: 616 LKVLDVSN 639
>AU251926
Length = 335
Score = 43.9 bits (102), Expect = 1e-04
Identities = 29/75 (38%), Positives = 41/75 (54%)
Frame = +2
Query: 658 LPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLK 717
LP +GKL +L L LS + A+P++IG L +L LD+ + + LP+ GNL NL
Sbjct: 107 LPDSLGKLSSLVTLDLSE-NRIVALPSTIGGLSSLTRLDL-HTNRIQELPDSIGNLLNLV 280
Query: 718 NLDMATCASIELPFS 732
LD+ LP S
Sbjct: 281 YLDLRGNQLPSLPAS 325
Score = 39.3 bits (90), Expect = 0.002
Identities = 24/59 (40%), Positives = 35/59 (58%)
Frame = +2
Query: 653 HKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFG 711
+++ +LP IG L +L L L + ++ +P SIG LLNL +LD+ L SLP FG
Sbjct: 161 NRIVALPSTIGGLSSLTRLDLHT-NRIQELPDSIGNLLNLVYLDLRG-NQLPSLPASFG 331
>BP063299
Length = 580
Score = 43.9 bits (102), Expect = 1e-04
Identities = 36/125 (28%), Positives = 58/125 (45%), Gaps = 2/125 (1%)
Frame = +3
Query: 264 LKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIA--TSLKNRPH 321
L+ L+ D LF YA N + +++V+ C GLPL + + LKN
Sbjct: 117 LQGLSENDCWLLFKQYAFGANNEERAELVAIGKEIVKKCGGLPLAARALGGLLRLKNEVK 296
Query: 322 DLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRIPVAA 381
+ WH+ VKE S ++ D L + F + P + +CF A+FP+D +I
Sbjct: 297 E-WHE-VKE-STLWNLPDERCILPALMLSYFHL---TPTLRQCFAFCAIFPKDAKIIKED 458
Query: 382 LVDMW 386
L+ +W
Sbjct: 459 LIYLW 473
>BG662453
Length = 460
Score = 43.5 bits (101), Expect = 1e-04
Identities = 55/172 (31%), Positives = 86/172 (49%), Gaps = 4/172 (2%)
Frame = +2
Query: 522 VEVLILNIHTKQY-SLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIR-LE 579
+ +++L++H+ Q SLP + +SKL+VL ++ N IE L +++E R LE
Sbjct: 38 LNMVVLDVHSNQLRSLPNSVGCLSKLKVLNVSG--------NLIEYLP--KSIENCRALE 187
Query: 580 RISVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGIC 639
++ N KLS + FE NL++L+++ K LV L
Sbjct: 188 ELNA-------NFNKLS--QLPDTMGFE---------LLNLKKLSVNSNK-LVFLPRSTS 310
Query: 640 DIISLKKLNVT-NCHKLSSLPQDIGKLENLELLSLS-SCTDLEAIPTSIGKL 689
+ SLK L+ NC L SLP D+ L NLE L++S + L+ +P SIG L
Sbjct: 311 HLTSLKILDARLNC--LRSLPDDLENLINLETLNVSQNFQYLDTLPYSIGLL 460
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.333 0.145 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,527,221
Number of Sequences: 28460
Number of extensions: 183971
Number of successful extensions: 1540
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 1458
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1511
length of query: 776
length of database: 4,897,600
effective HSP length: 97
effective length of query: 679
effective length of database: 2,136,980
effective search space: 1451009420
effective search space used: 1451009420
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148347.4


