

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
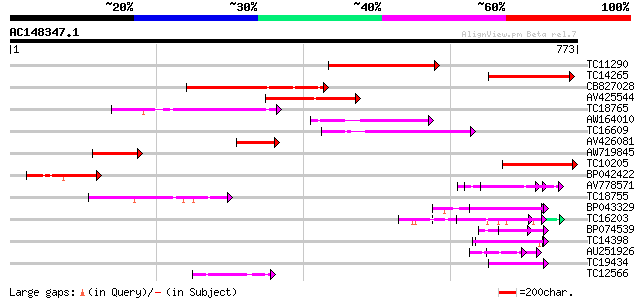
Query= AC148347.1 + phase: 0
(773 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC11290 weakly similar to UP|AAN62760 (AAN62760) Disease resista... 193 1e-49
TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resista... 187 4e-48
CB827028 154 7e-38
AV425544 140 8e-34
TC18765 weakly similar to UP|Q9LVT1 (Q9LVT1) Disease resistance ... 127 5e-30
AW164010 119 2e-27
TC16609 weakly similar to PIR|T48468|T48468 disease resistance-l... 107 7e-24
AV426081 103 1e-22
AW719845 98 4e-21
TC10205 weakly similar to UP|Q8RL77 (Q8RL77) HpaG, partial (10%) 87 8e-18
BP042422 80 2e-15
AV778571 62 3e-10
TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fr... 56 2e-08
BP043329 55 6e-08
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 54 7e-08
BP074539 47 9e-06
TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%) 46 3e-05
AU251926 45 4e-05
TC19434 similar to UP|Q94F63 (Q94F63) Somatic embryogenesis rece... 45 4e-05
TC12566 similar to UP|Q84XI1 (Q84XI1) RCa1 (Fragment), partial (... 45 6e-05
>TC11290 weakly similar to UP|AAN62760 (AAN62760) Disease resistance
protein-like protein MsR1, partial (18%)
Length = 459
Score = 193 bits (490), Expect = 1e-49
Identities = 103/152 (67%), Positives = 120/152 (78%)
Frame = +3
Query: 435 GNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQGTMAHILSKLIGWFDKPKPQKVPART 494
G YQ+ QEP+ +RKR I+TNE K + L+EKQQG M+ LSK+ K KPQ V ART
Sbjct: 3 GIYQSIQEPVGKRKRLNIDTNEDKLEWLLQEKQQGIMSRTLSKIRRLCLKQKPQLVLART 182
Query: 495 LSISIDETCASDWSQVQPALAEVLILNLQTKQYTFPELMEKMNKLKALIVINHGLRPSEL 554
LSIS DETC D S +QPA AEVLILNL+TK+Y+ PE++E+MNKLKALIV N+G PSEL
Sbjct: 183 LSISTDETCTPDLSLIQPAEAEVLILNLRTKKYSLPEILEEMNKLKALIVTNYGFHPSEL 362
Query: 555 NNLELLSSLSNLKRIRLERISVPSFGTLKNLK 586
NN ELL SLSNLKRIRLE+ISVPSFGTLKNLK
Sbjct: 363 NNFELLDSLSNLKRIRLEQISVPSFGTLKNLK 458
>TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resistance
protein-like protein MsR1, partial (5%)
Length = 766
Score = 187 bits (476), Expect = 4e-48
Identities = 92/118 (77%), Positives = 102/118 (85%)
Frame = +1
Query: 653 QEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNL 712
QEIGKLENLELL L SCTDL LPDSIGRLS LRLLDISNCISL SLPE+ GNLCNL++L
Sbjct: 1 QEIGKLENLELLRLSSCTDLKGLPDSIGRLSKLRLLDISNCISLPSLPEEIGNLCNLKSL 180
Query: 713 DMTSCASCELPFSVVNLQNLKVTCDEETAASWESFQSMISNLTIEVPHVEVNLNWLHA 770
MTSCA CELP S+VNLQNL V CDEETAASWE+F+ +I +L IEVP V+VNLNWLH+
Sbjct: 181 YMTSCAGCELPSSIVNLQNLTVVCDEETAASWEAFEYVIPHLKIEVPQVDVNLNWLHS 354
>CB827028
Length = 572
Score = 154 bits (388), Expect = 7e-38
Identities = 88/195 (45%), Positives = 129/195 (66%), Gaps = 2/195 (1%)
Frame = +2
Query: 242 SRVAFSRFDK--TFILKPLAQEDSVTLFRHYTEVEKNSSKIPDKDLIEKVVEHCKGLPLA 299
SR A +R+D ++L+ L+ + + +LF H + NS++IP L++K+V+ CK P+A
Sbjct: 2 SRSAIARYDSGSPYVLERLSLDYATSLFLHSASLSPNSTEIP-VSLVKKIVKGCKRSPIA 178
Query: 300 IKVIATSFRYRPYELWEKIVKELSRGRSILDSNTELLIRLQKILDVLEDNAISKECFMDL 359
+ V TS + + +W K LS+G+SIL+SN ++L LQK LDVL+ AI ECF DL
Sbjct: 179 LTVTGTSLKNKNLAVWRSRAKALSKGQSILESNHDVLTCLQKSLDVLDPIAI--ECFKDL 352
Query: 360 ALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIARKNASDTENYY 419
LFPEDQRIP AAL+DMWAEL DD+ AM+ I +L ++N+A++ + RK A+D +Y
Sbjct: 353 GLFPEDQRIPAAALVDMWAELRCGDDE--TAMEKIYELVNLNMADIFVTRKVANDIVDYN 526
Query: 420 YNNHFIVLHDLLREL 434
Y H++ H LLREL
Sbjct: 527 Y--HYVAQHGLLREL 565
>AV425544
Length = 425
Score = 140 bits (353), Expect = 8e-34
Identities = 73/131 (55%), Positives = 92/131 (69%), Gaps = 1/131 (0%)
Frame = +1
Query: 349 NAISKECFMDLALFPEDQRIPVAALIDMWAELYGLDDDGKEAMDIINKLDSMNLANVLIA 408
N KECFMDL LFPEDQRIPV ALIDMWAELY LD+DG+ AM I+ L S NL N ++
Sbjct: 4 NINEKECFMDLGLFPEDQRIPVTALIDMWAELYNLDEDGRNAMTIVLDLTSRNLINFIVT 183
Query: 409 RKNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTN-ESKCDQRLREKQ 467
RK ASD YNNHF++LHDLLREL +Q+ EP EQRKR +I+ N +++ + + + Q
Sbjct: 184 RKVASDA-GVCYNNHFVMLHDLLRELAIHQSKGEPFEQRKRLIIDLNGDNRPEWWVGQNQ 360
Query: 468 QGTMAHILSKL 478
QG + + S L
Sbjct: 361 QGFIGRLFSFL 393
>TC18765 weakly similar to UP|Q9LVT1 (Q9LVT1) Disease resistance
protein-like, partial (18%)
Length = 688
Score = 127 bits (320), Expect = 5e-30
Identities = 85/242 (35%), Positives = 136/242 (56%), Gaps = 10/242 (4%)
Frame = +2
Query: 139 KTTLATKLCLDDQVKGKFKENIIFVTFSKTPMLKIIVERLFEH--------CGYPVPEYQ 190
KTTLA ++C DDQV+ FKE I+F+T S++P ++ + ++F H Y VP++
Sbjct: 2 KTTLAREVCRDDQVRCHFKERILFLTVSQSPNVEELRAKIFGHIMGNRGLNANYAVPQWM 181
Query: 191 SDEDAVNGLGLLLRKIEGSPILLVLDDVWPGSEDLVEKFKFQISDYKILVTSRVAFSR-F 249
+ + S IL+VLDDVW S ++E+ ++ K LV SR F R F
Sbjct: 182 PQFECQSQ----------SQILVVLDDVW--SLPVLEQLVLRVPGCKYLVVSRFKFQRIF 325
Query: 250 DKTFILKPLAQEDSVTLFRHYTEVEKNSSKIPDKDLIEKVVEHCKGLPLAIKVIATSFRY 309
+ T+ ++ L++ D+++LF H+ K+ +++LI++VV C LPLA+KVI S R
Sbjct: 326 NDTYDVELLSEGDALSLFCHHAFGHKSIPFGANQNLIKQVVAECGRLPLALKVIGASLRD 505
Query: 310 RPYELWEKIVKELSRGRSILDS-NTELLIRLQKILDVLEDNAISKECFMDLALFPEDQRI 368
+ W + LS+G SI +S L+ R+ + L + KECF+DL FPED++I
Sbjct: 506 QNEMFWLSVKTRLSQGLSIGESYEVNLIDRMAISTNYLPEKV--KECFLDLCAFPEDKKI 679
Query: 369 PV 370
P+
Sbjct: 680 PL 685
>AW164010
Length = 494
Score = 119 bits (297), Expect = 2e-27
Identities = 67/168 (39%), Positives = 93/168 (54%)
Frame = +2
Query: 410 KNASDTENYYYNNHFIVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQG 469
K ASD +Y Y H++ H LLR+L Q ++E ++R R +I N +
Sbjct: 17 KVASDIVDYNY--HYVTQHGLLRDLAILQTSEETEDKRDRLIIEINRNN----------- 157
Query: 470 TMAHILSKLIGWFDKPKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTF 529
L W+ + AR LSIS DE +S+ +QP E L+LNL+ K+YT
Sbjct: 158 --------LPAWWSLENEYPIAARILSISTDEAFSSELCNLQPTKVEALVLNLRGKKYTL 313
Query: 530 PELMEKMNKLKALIVINHGLRPSELNNLELLSSLSNLKRIRLERISVP 577
P MEKMNKLK L + N+ L P+EL N E+L LSNLKR+RLE++ +P
Sbjct: 314 PMFMEKMNKLKVLTITNYDLYPAELENFEMLDHLSNLKRLRLEKVRIP 457
>TC16609 weakly similar to PIR|T48468|T48468 disease resistance-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(12%)
Length = 621
Score = 107 bits (267), Expect = 7e-24
Identities = 64/212 (30%), Positives = 110/212 (51%), Gaps = 2/212 (0%)
Frame = +1
Query: 425 IVLHDLLRELGNYQNTQEPIEQRKRQLINTNESKCDQRLREKQQGTMAHILSKLIGWFDK 484
+ HD+LR+L + + I +R R ++ E Q +E W +
Sbjct: 34 VTQHDILRDLALNLSNRGSINERLRLVMPKREGN-GQLPKE---------------WL-R 162
Query: 485 PKPQKVPARTLSISIDETCASDWSQVQPALAEVLILNLQTKQYTFPELMEKMNKLKALIV 544
+ Q + AR +SI E DW +++ AEVLILN + +Y P + +M L+ALIV
Sbjct: 163 YRGQPLEARIVSIHTGEMTEGDWCELEFPKAEVLILNFTSSEYFLPPFIARMPSLRALIV 342
Query: 545 INHGLRPSELNNLELLSSLSNLKRIRLERISVPSFG--TLKNLKKLSLYMCNTRLAFEKG 602
IN+ + L+N+ + +LSNL+ + LE++S P L+ L KL + +C + +
Sbjct: 343 INYSTTYAHLHNVSVFKNLSNLRSLWLEKVSTPQLSGIVLEKLGKLFIVLCQVNNSLKGK 522
Query: 603 SILISDLFPNLEDLSMDYCKDMTALPNGVCDI 634
++ +FPNL + ++D+C D+T LP+ +C I
Sbjct: 523 DANLAHIFPNLSEFTLDHCDDLTELPSSICGI 618
>AV426081
Length = 179
Score = 103 bits (257), Expect = 1e-22
Identities = 49/59 (83%), Positives = 54/59 (91%)
Frame = +1
Query: 310 RPYELWEKIVKELSRGRSILDSNTELLIRLQKILDVLEDNAISKECFMDLALFPEDQRI 368
RPYELW+KIVKELS+G S+LDS+TELL RLQKILDV EDN + KECFMDLALFPEDQRI
Sbjct: 1 RPYELWQKIVKELSQGHSVLDSHTELLTRLQKILDVAEDNPVIKECFMDLALFPEDQRI 177
>AW719845
Length = 209
Score = 98.2 bits (243), Expect = 4e-21
Identities = 46/68 (67%), Positives = 58/68 (84%)
Frame = +3
Query: 114 LIKLKVEILREGRSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTFSKTPMLKI 173
L KLK+E+L++G S + +TGLGG+GKTTLA +C D Q+KGKF+ENI+FVTFSKTP +K
Sbjct: 3 LSKLKMELLKDGVSVVAVTGLGGLGKTTLAAMVCWDKQIKGKFRENILFVTFSKTPNVKN 182
Query: 174 IVERLFEH 181
IVERLFEH
Sbjct: 183 IVERLFEH 206
>TC10205 weakly similar to UP|Q8RL77 (Q8RL77) HpaG, partial (10%)
Length = 586
Score = 87.4 bits (215), Expect = 8e-18
Identities = 48/103 (46%), Positives = 67/103 (64%), Gaps = 2/103 (1%)
Frame = +3
Query: 673 VELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCAS-CELPFSVVNLQN 731
+ELP SI L ++ LDIS+CISLS LPED G L L L++ C+ ELP S+++L+N
Sbjct: 3 LELPKSITNLEQVQFLDISDCISLSHLPEDMGELKKLEKLNVRGCSRLTELPTSIMDLEN 182
Query: 732 L-KVTCDEETAASWESFQSMISNLTIEVPHVEVNLNWLHANRS 773
L +V CDEE A WE Q ++S+L + V + NL++L RS
Sbjct: 183 LEEVECDEEIAELWEPVQQILSDLQLHVAPTDFNLDFLSKLRS 311
Score = 36.2 bits (82), Expect = 0.020
Identities = 27/81 (33%), Positives = 46/81 (56%), Gaps = 5/81 (6%)
Frame = +3
Query: 612 NLED---LSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLIS 668
NLE L + C ++ LP + ++ L+KL++ C +L+ LP I LENLE +
Sbjct: 27 NLEQVQFLDISDCISLSHLPEDMGELKKLEKLNVRGCSRLTELPTSIMDLENLEE---VE 197
Query: 669 C-TDLVELPDSIGR-LSNLRL 687
C ++ EL + + + LS+L+L
Sbjct: 198 CDEEIAELWEPVQQILSDLQL 260
>BP042422
Length = 526
Score = 79.7 bits (195), Expect = 2e-15
Identities = 45/105 (42%), Positives = 70/105 (65%), Gaps = 3/105 (2%)
Frame = +3
Query: 24 KNLCCIKWFSLVKRCIFVVFYKKKNKDDFDSCVGDDDDKQKMGKDVEG---KLRELLKIL 80
K LC + SL++ C+ + + NKDDF + D + + +M DV+ K+R L+++
Sbjct: 222 KCLCWGNFISLLQLCVNKLCHN--NKDDFLNV--DGEKQAQMANDVKDTLYKVRGFLELI 389
Query: 81 DKENFGKKISGSILKGPFDVPANPEFTVGLDLQLIKLKVEILREG 125
KE+F +K SG+ +K P+ VPANPEFTVG+D+ L KLKVE+L++G
Sbjct: 390 SKEDFEQKFSGAPIKRPYGVPANPEFTVGVDVPLNKLKVELLKDG 524
>AV778571
Length = 452
Score = 62.0 bits (149), Expect = 3e-10
Identities = 36/113 (31%), Positives = 57/113 (49%)
Frame = +3
Query: 611 PNLEDLSMDYCKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCT 670
PNLE LS Y +++ + V + L+ L I C K+ P KL +LE L L C+
Sbjct: 84 PNLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPM--KLASLEELYLSHCS 257
Query: 671 DLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCASCELP 723
L P+ + ++ N+ L+ + + + LP GNL LR L++ C +LP
Sbjct: 258 SLESFPEILAKMENITLIGVYHT-PIKELPSSIGNLTRLRRLELHGCGMLQLP 413
Score = 50.8 bits (120), Expect = 8e-07
Identities = 33/114 (28%), Positives = 60/114 (51%), Gaps = 1/114 (0%)
Frame = +3
Query: 643 TNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPED 702
+ C ++ +P ++ L NLE LS + +L+++ +S+G L+ LR+LDI C + + P
Sbjct: 39 SECESITQIP-DLSGLPNLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPP- 212
Query: 703 FGNLCNLRNLDMTSCASCE-LPFSVVNLQNLKVTCDEETAASWESFQSMISNLT 755
L +L L ++ C+S E P + ++N+ + T + S I NLT
Sbjct: 213 -MKLASLEELYLSHCSSLESFPEILAKMENITLIGVYHTPI--KELPSSIGNLT 365
Score = 49.7 bits (117), Expect = 2e-06
Identities = 32/113 (28%), Positives = 57/113 (50%)
Frame = +3
Query: 621 CKDMTALPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIG 680
C+ +T +P+ + + +L+KLS L + + +G L L +L + C + P
Sbjct: 45 CESITQIPD-LSGLPNLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPM-- 215
Query: 681 RLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCASCELPFSVVNLQNLK 733
+L++L L +S+C SL S PE + N+ + + ELP S+ NL L+
Sbjct: 216 KLASLEELYLSHCSSLESFPEILAKMENITLIGVYHTPIKELPSSIGNLTRLR 374
>TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fragment),
complete
Length = 880
Score = 55.8 bits (133), Expect = 2e-08
Identities = 60/215 (27%), Positives = 104/215 (47%), Gaps = 18/215 (8%)
Frame = +3
Query: 108 VGLDLQLIKLKVEILREG--RSTLLLTGLGGMGKTTLATKLCLDDQVKGKFKENIIFVTF 165
VG+D + KL +++ R + +TG+GGMGKTTL K DD V K ++T
Sbjct: 198 VGIDRRKKKLMGCLIKPCPVRKVISVTGMGGMGKTTL-VKQVYDDPVVIKHFRACAWITV 374
Query: 166 SKT----PMLKIIVERLFEHCGYPVPEYQSDEDAVNGLGLLLRK-IEGSPILLVLDDVWP 220
S++ +L+ + +LF PVP + + L ++++ ++ L+V DDVW
Sbjct: 375 SQSCEIGELLRDLARQLFSEIRRPVP-LGLENMRCDRLKMIIKDLLQRRRYLVVFDDVWH 551
Query: 221 GSEDLVEKFKFQISD----YKILVTSRVAFSRF-------DKTFILKPLAQEDSVTLFRH 269
E E K+ + D +I++T+R + F K + L+PL ++++ LF
Sbjct: 552 VRE--WEAVKYALPDNNCGSRIMITTRRSDLAFTSSTESKGKVYNLQPLKEDEAWELFCR 725
Query: 270 YTEVEKNSSKIPDKDLIEKVVEHCKGLPLAIKVIA 304
T +S + ++ C+GLPLAI I+
Sbjct: 726 KT-FHGDSCPSHLIGICTYILRKCEGLPLAIVAIS 827
>BP043329
Length = 484
Score = 54.7 bits (130), Expect = 6e-08
Identities = 36/110 (32%), Positives = 60/110 (53%), Gaps = 2/110 (1%)
Frame = +3
Query: 627 LPNGVCDIISLKKLSIT-NCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNL 685
+P + + +LK L+++ N +P EIG L NLE++ L C + +PDSIG L L
Sbjct: 6 IPPSLGTLTTLKMLNLSYNPFYPGRIPPEIGNLTNLEVVWLTQCNLVGVIPDSIGNLKKL 185
Query: 686 RLLDISNCISLSSLPEDFGNLCNLRNLDM-TSCASCELPFSVVNLQNLKV 734
+ LD++ S+P L +LR +++ + S ELP + NL L++
Sbjct: 186 KDLDLALNDLYGSIPSSLTGLTSLRQIELYNNSLSGELPRGMGNLTELRL 335
Score = 48.5 bits (114), Expect = 4e-06
Identities = 46/160 (28%), Positives = 76/160 (46%), Gaps = 4/160 (2%)
Frame = +3
Query: 577 PSFGTLKNLKKLSLY---MCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCD 633
PS GTL LK L+L R+ E G++ NLE + + C + +P+ + +
Sbjct: 12 PSLGTLTTLKMLNLSYNPFYPGRIPPEIGNLT------NLEVVWLTQCNLVGVIPDSIGN 173
Query: 634 IISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNC 693
+ LK L + +P + L +L + L + + ELP +G L+ LRLLD S
Sbjct: 174 LKKLKDLDLALNDLYGSIPSSLTGLTSLRQIELYNNSLSGELPRGMGNLTELRLLDASMN 353
Query: 694 ISLSSLPEDFGNLCNLRNLDM-TSCASCELPFSVVNLQNL 732
+PE+ +L L +L++ + ELP S+ + NL
Sbjct: 354 HLTGRIPEELCSL-PLESLNLYENRFEGELPASIADSPNL 470
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation
aberrant root formation protein), complete
Length = 3308
Score = 54.3 bits (129), Expect = 7e-08
Identities = 58/192 (30%), Positives = 84/192 (43%), Gaps = 7/192 (3%)
Frame = +2
Query: 530 PELMEKMNKLKALIVINH---GLRP---SELNNLELLSSLSNLKRIRLERISVPSFGTLK 583
PE + K+ KLK L + + G P SE +LE L +N R+ S LK
Sbjct: 599 PEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPE----SLAKLK 766
Query: 584 NLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIISLKKLSIT 643
LK+L L N A+E G NL L M C +P + ++ L L +
Sbjct: 767 TLKELHLGYSN---AYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQ 937
Query: 644 NCHKLSLLPQEIGKLENLELLSLISCTDLV-ELPDSIGRLSNLRLLDISNCISLSSLPED 702
+ +P E+ + +L L L S DL E+P+S +L NL L++ SLP
Sbjct: 938 MNNLTGTIPPELSSMMSLMSLDL-SINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSF 1114
Query: 703 FGNLCNLRNLDM 714
G+L NL L +
Sbjct: 1115IGDLPNLETLQV 1150
Score = 49.7 bits (117), Expect = 2e-06
Identities = 34/126 (26%), Positives = 67/126 (52%), Gaps = 3/126 (2%)
Frame = +2
Query: 610 FPNLEDLSMDYCKDMTALPNGVCDIISLKKLSI--TNCHKLSLLPQEIGKLENLELLSLI 667
F +LE L ++ +P + + +LK+L + +N ++ + P G +ENL LL +
Sbjct: 689 FQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGI-PPAFGSMENLRLLEMA 865
Query: 668 SCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMT-SCASCELPFSV 726
+C E+P S+G L+ L L + ++P + ++ +L +LD++ + + E+P S
Sbjct: 866 NCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESF 1045
Query: 727 VNLQNL 732
L+NL
Sbjct: 1046SKLKNL 1063
Score = 40.8 bits (94), Expect = 8e-04
Identities = 59/237 (24%), Positives = 96/237 (39%), Gaps = 57/237 (24%)
Frame = +2
Query: 577 PSFGTLKNLKKLSLYMCNTRLAFEKGSILISDLFPNLEDLSMDYCKDMTALPNGVCDIIS 636
P G L+ L+ L++ M N L + S L S +L+ L++ + P + +
Sbjct: 383 PEIGLLEKLENLTISMNN--LTDQLPSDLAS--LTSLKVLNISHNLFSGQFPGNIT--VG 544
Query: 637 LKKLSITNCHKLSL---LPQEIGKLENLELL----------------------------- 664
+ +L + + S LP+EI KLE L+ L
Sbjct: 545 MTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNAN 724
Query: 665 --------SLISCTDLVEL------------PDSIGRLSNLRLLDISNCISLSSLPEDFG 704
SL L EL P + G + NLRLL+++NC +P G
Sbjct: 725 SLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLG 904
Query: 705 NLCNLRNL-----DMTSCASCELPFSVVNLQNLKVTCDEETAASWESFQSMISNLTI 756
NL L +L ++T EL S+++L +L ++ ++ T ESF S + NLT+
Sbjct: 905 NLTKLHSLFVQMNNLTGTIPPELS-SMMSLMSLDLSINDLTGEIPESF-SKLKNLTL 1069
Score = 38.5 bits (88), Expect = 0.004
Identities = 30/96 (31%), Positives = 48/96 (49%), Gaps = 2/96 (2%)
Frame = +2
Query: 640 LSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSL 699
L++T LP EIG LE LE L++ +LP + L++L++L+IS+ +
Sbjct: 344 LNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQF 523
Query: 700 PEDFG-NLCNLRNLD-MTSCASCELPFSVVNLQNLK 733
P + + L LD + S LP +V L+ LK
Sbjct: 524 PGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLK 631
Score = 27.7 bits (60), Expect = 7.3
Identities = 23/100 (23%), Positives = 48/100 (48%), Gaps = 1/100 (1%)
Frame = +2
Query: 636 SLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCIS 695
SL L+++N +P + L L+ LSL + + E+P + + L ++IS
Sbjct: 1487 SLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNL 1666
Query: 696 LSSLPEDFGNLCNLRNLDMT-SCASCELPFSVVNLQNLKV 734
+P + +L +D++ + + E+P + NL +L +
Sbjct: 1667 TGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSI 1786
>BP074539
Length = 357
Score = 47.4 bits (111), Expect = 9e-06
Identities = 26/68 (38%), Positives = 39/68 (57%)
Frame = +1
Query: 667 ISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLDMTSCASCELPFSV 726
+S T++ +LPDS L NL++L + NC L LP + L NLR LD + ++P
Sbjct: 28 LSHTNIEKLPDSTCLLYNLQILKLKNCRYLKELPLNLHKLTNLRYLDFSGTQVRKMPMHF 207
Query: 727 VNLQNLKV 734
L+NL+V
Sbjct: 208 GKLKNLQV 231
Score = 45.8 bits (107), Expect = 3e-05
Identities = 31/74 (41%), Positives = 42/74 (55%)
Frame = +1
Query: 640 LSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSL 699
LS TN KL P L NL++L L +C L ELP ++ +L+NLR LD S + +
Sbjct: 28 LSHTNIEKL---PDSTCLLYNLQILKLKNCRYLKELPLNLHKLTNLRYLDFSG-TQVRKM 195
Query: 700 PEDFGNLCNLRNLD 713
P FG L NL+ L+
Sbjct: 196 PMHFGKLKNLQVLN 237
>TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%)
Length = 982
Score = 45.8 bits (107), Expect = 3e-05
Identities = 30/103 (29%), Positives = 50/103 (48%), Gaps = 5/103 (4%)
Frame = +1
Query: 636 SLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCIS 695
SL+ L + + +P+E+G L++L L L +P S+G L NLR L ++N
Sbjct: 397 SLQYLELYENNIQGTIPEELGNLQSLISLDLYHNNVSGSIPSSLGNLKNLRFLRLNNNHL 576
Query: 696 LSSLPEDFGNLCNLRNLDMTSCASC-----ELPFSVVNLQNLK 733
+P+ L NL+ LD+++ C PF + L N +
Sbjct: 577 TGQIPKSLSTLPNLKVLDVSNNNLCGPIPTSGPFEHIPLDNFE 705
Score = 45.8 bits (107), Expect = 3e-05
Identities = 31/104 (29%), Positives = 53/104 (50%), Gaps = 1/104 (0%)
Frame = +1
Query: 632 CDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDIS 691
C S+ ++ + N + L ++G L +L+ L L +P+ +G L +L LD+
Sbjct: 313 CQDNSVTRVDLGNLNLSGHLVPDLGNLHSLQYLELYENNIQGTIPEELGNLQSLISLDLY 492
Query: 692 NCISLSSLPEDFGNLCNLRNLDM-TSCASCELPFSVVNLQNLKV 734
+ S+P GNL NLR L + + + ++P S+ L NLKV
Sbjct: 493 HNNVSGSIPSSLGNLKNLRFLRLNNNHLTGQIPKSLSTLPNLKV 624
>AU251926
Length = 335
Score = 45.1 bits (105), Expect = 4e-05
Identities = 31/75 (41%), Positives = 41/75 (54%)
Frame = +2
Query: 651 LPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLR 710
LP +GKL +L L L S +V LP +IG LS+L LD+ + + LP+ GNL NL
Sbjct: 107 LPDSLGKLSSLVTLDL-SENRIVALPSTIGGLSSLTRLDL-HTNRIQELPDSIGNLLNLV 280
Query: 711 NLDMTSCASCELPFS 725
LD+ LP S
Sbjct: 281 YLDLRGNQLPSLPAS 325
Score = 41.6 bits (96), Expect = 5e-04
Identities = 31/78 (39%), Positives = 43/78 (54%)
Frame = +2
Query: 627 LPNGVCDIISLKKLSITNCHKLSLLPQEIGKLENLELLSLISCTDLVELPDSIGRLSNLR 686
LP+ + + SL L ++ +++ LP IG L +L L L + + ELPDSIG L NL
Sbjct: 107 LPDSLGKLSSLVTLDLSE-NRIVALPSTIGGLSSLTRLDLHT-NRIQELPDSIGNLLNLV 280
Query: 687 LLDISNCISLSSLPEDFG 704
LD+ L SLP FG
Sbjct: 281 YLDLRG-NQLPSLPASFG 331
>TC19434 similar to UP|Q94F63 (Q94F63) Somatic embryogenesis receptor-like
kinase 2, partial (26%)
Length = 550
Score = 45.1 bits (105), Expect = 4e-05
Identities = 27/82 (32%), Positives = 46/82 (55%), Gaps = 1/82 (1%)
Frame = +2
Query: 654 EIGKLENLELLSLISCTDLVELPDSIGRLSNLRLLDISNCISLSSLPEDFGNLCNLRNLD 713
++G+L NL+ L L S ++PD +G L+NL LD+ ++P GNL LR L
Sbjct: 29 QLGQLSNLQYLELYSNNMTGKIPDELGNLTNLVSLDLYLNNLTGNIPSTLGNLGKLRFLR 208
Query: 714 M-TSCASCELPFSVVNLQNLKV 734
+ + S +P ++ N+ +L+V
Sbjct: 209 LNNNTLSGGIPMTLTNVSSLQV 274
>TC12566 similar to UP|Q84XI1 (Q84XI1) RCa1 (Fragment), partial (28%)
Length = 918
Score = 44.7 bits (104), Expect = 6e-05
Identities = 32/113 (28%), Positives = 60/113 (52%)
Frame = +1
Query: 250 DKTFILKPLAQEDSVTLFRHYTEVEKNSSKIPDKDLIEKVVEHCKGLPLAIKVIATSFRY 309
D+ + +K ++ EDS+ LF ++N DL +K++ + KG+PLA+KV+
Sbjct: 4 DEIYEVKEMSFEDSLQLFS-LNAFKRNDPLETYIDLSKKLLSYAKGIPLALKVLGLFLYG 180
Query: 310 RPYELWEKIVKELSRGRSILDSNTELLIRLQKILDVLEDNAISKECFMDLALF 362
R + WE + +L + + E++ L+ D L+D+ K+ F+D+A F
Sbjct: 181 RERKAWESQLVKLEK-----LPDPEIINVLKLSYDGLDDD--QKDIFLDIACF 318
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.138 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,937,626
Number of Sequences: 28460
Number of extensions: 176278
Number of successful extensions: 931
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 860
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 899
length of query: 773
length of database: 4,897,600
effective HSP length: 97
effective length of query: 676
effective length of database: 2,136,980
effective search space: 1444598480
effective search space used: 1444598480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148347.1


