

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.6 + phase: 0
(570 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
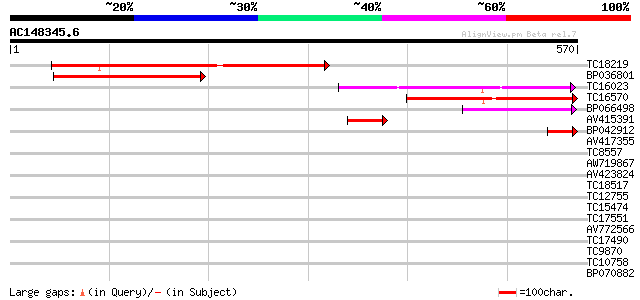
Score E
Sequences producing significant alignments: (bits) Value
TC18219 271 3e-73
BP036801 228 3e-60
TC16023 180 6e-46
TC16570 similar to UP|Q9LR03 (Q9LR03) F10A5.18, partial (18%) 166 7e-42
BP066498 86 2e-17
AV415391 64 5e-11
BP042912 58 5e-09
AV417355 33 0.16
TC8557 similar to UP|Q9LK52 (Q9LK52) Dbj|BAA90629.1, complete 31 0.47
AW719867 30 1.1
AV423824 30 1.1
TC18517 weakly similar to UP|AAQ82688 (AAQ82688) Epa4p, partial ... 30 1.4
TC12755 similar to UP|Q9C587 (Q9C587) Replication factor C large... 30 1.4
TC15474 UP|Q7Y1B9 (Q7Y1B9) Ammonium transporter, complete 30 1.4
TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, part... 29 1.8
AV772566 29 1.8
TC17490 similar to GB|AAO64817.1|29028876|BT005882 At3g04570 {Ar... 29 1.8
TC9870 weakly similar to UP|NO16_MEDTR (P93328) Early nodulin 16... 29 2.4
TC10758 29 2.4
BP070882 29 2.4
>TC18219
Length = 936
Score = 271 bits (692), Expect = 3e-73
Identities = 136/286 (47%), Positives = 182/286 (63%), Gaps = 7/286 (2%)
Frame = +2
Query: 43 SPSSNQGEWELIQPTIGISAMHMQLSHNNKIIIFDRTDFGPSNLPL----SNGRCRMDPF 98
S +G+W+++ G+ MH L+ N II+FD+T+ GPS PL + RC
Sbjct: 89 SLEGRKGQWQMLLNNTGVVGMHTALTSKNTIIMFDQTEAGPSGYPLHKRFNGSRCNTRSH 268
Query: 99 DTALKIDCTAHSVLYDIATNTFRSLTVQTDTWCSSGSVLSNGTLVQTGGFNDGERRIRMF 158
+ + C AHSV YDI+ N R L + +D WCSSGS LSNGTL+QTGG+ G +RIR +
Sbjct: 269 NDVVDSTCYAHSVEYDISANRVRPLRLDSDPWCSSGSFLSNGTLLQTGGYGRGAKRIRFY 448
Query: 159 TPCFNENCDWIEFPSYLSERRWYATNQILPD-NRIIIIGGRRQFNYEFIPKTTTSSSSSS 217
PC N C WI+ LS+ RWYA++QILPD +R++I+GGRR F YEFIPK S +
Sbjct: 449 RPCGNHQCHWIQSRKSLSDERWYASSQILPDHDRVVIVGGRRTFTYEFIPKI-----SPT 613
Query: 218 SSSIHLSFLQETNDPSE--NNLYPFVHLLPNGNLFIFANTRSILFDYKQNVVVKEFPEIP 275
S L FL +TN+ + NNLYPF+HL +GNLFIFAN SIL + ++N V+K FP IP
Sbjct: 614 EKSFDLPFLHKTNERNAKGNNLYPFLHLSSDGNLFIFANRDSILLNPRRNRVIKTFPRIP 793
Query: 276 GGDPHNYPSSGSSVLLPLDENQISMEATIMICGGAPRGSFEAAKGK 321
G NYPSSGSSV+LPLD + + +M+CGG+ G+ AA +
Sbjct: 794 GDGSRNYPSSGSSVMLPLDHSDNFHKVEVMVCGGSATGALRAASNR 931
>BP036801
Length = 540
Score = 228 bits (580), Expect = 3e-60
Identities = 102/154 (66%), Positives = 125/154 (80%), Gaps = 1/154 (0%)
Frame = +1
Query: 45 SSNQGEWELIQPTIGISAMHMQLSHNNKIIIFDRTDFGPSNLPLSNGRCRMDPFDTALKI 104
SS G W +Q +IGISAMHMQ+ ++NK++IFDRTDFGPSN+ LSNGRCR +P D ALK+
Sbjct: 79 SSTGGHWVQLQRSIGISAMHMQVMNDNKVVIFDRTDFGPSNISLSNGRCRFNPRDMALKL 258
Query: 105 DCTAHSVLYDIATNTFRSLTVQTDTWCSSGSVLSNGTLVQTGGFNDGERRIRMFTPCFNE 164
DCTAHSVLYD+ TNTFR LT+QTD WCSSG++ +GTL+QTGGFNDG ++R FTPC
Sbjct: 259 DCTAHSVLYDLTTNTFRPLTIQTDAWCSSGALTPDGTLLQTGGFNDGYHKLRTFTPCPQH 438
Query: 165 N-CDWIEFPSYLSERRWYATNQILPDNRIIIIGG 197
N CDW+E P L+ RWYA+NQILPD RII++GG
Sbjct: 439 NLCDWLELPQNLTSPRWYASNQILPDGRIIVVGG 540
>TC16023
Length = 894
Score = 180 bits (456), Expect = 6e-46
Identities = 99/244 (40%), Positives = 144/244 (58%), Gaps = 6/244 (2%)
Frame = +2
Query: 331 GFLKVTDSNPSWIIENMPMARVMGDMLILPNGDVIIINGAGSGTAGWENGRQPVLTPVIF 390
G + +T ++ W +E MP R++ DMLILPNG+++IINGA G AG++N R L P ++
Sbjct: 2 GRMVITXNDNKWEMEYMPEPRLLHDMLILPNGNILIINGAKHGCAGYDNARNASLEPYLY 181
Query: 391 RSSETKSDKRFSVMSPASRPRLYHSSAIVLRDGRVLVGGSNPHVNYNFTGVEFPTDLSLE 450
S + + KRF V+ R+YHSSA +L DGRVLV G NPH Y+F V +PT+L L+
Sbjct: 182 -SPKKRLGKRFRVLKSTKIARMYHSSATLLPDGRVLVAGGNPHGRYSFHNVAYPTELRLQ 358
Query: 451 AFSPPYLSLEFDLVRPTIWHVTN------KILGYRVFYYVTFTVAKFASASEVSVRLLAP 504
AF P Y+ + RP+ V + ++GY + V F V S +EV+ + AP
Sbjct: 359 AFVPHYMHRRYHSWRPSNLTVKSGGGGERGVIGYGAEFRVRFLVGTRPS-NEVAFSVYAP 535
Query: 505 SFTTHSFGMNQRMVVLKLIGVTMVNLDIYYATVVGPSTQEIAPPGYYLLFLVHAGVPSSG 564
FTTHS MNQRM+ L+ + + A + P + +AP GYYLL +V+ G+PS
Sbjct: 536 PFTTHSVAMNQRMLKLRCRSMVRSSGGWVDAVLEAPPSPRVAPSGYYLLTVVNGGIPSVS 715
Query: 565 EWVQ 568
+W+Q
Sbjct: 716 QWIQ 727
Score = 26.9 bits (58), Expect = 8.9
Identities = 9/28 (32%), Positives = 17/28 (60%)
Frame = +2
Query: 178 RRWYATNQILPDNRIIIIGGRRQFNYEF 205
R ++++ +LPD R+++ GG Y F
Sbjct: 239 RMYHSSATLLPDGRVLVAGGNPHGRYSF 322
>TC16570 similar to UP|Q9LR03 (Q9LR03) F10A5.18, partial (18%)
Length = 794
Score = 166 bits (421), Expect = 7e-42
Identities = 86/178 (48%), Positives = 116/178 (64%), Gaps = 7/178 (3%)
Frame = +3
Query: 400 RFSVMSPASRPRLYHSSAIVLRDGRVLVGGSNPHVNYNFTGVEFPTDLSLEAFSPPYLSL 459
+F + +P P +YHS+A++LRDGRVLV GSNPH YNF+ V FPT+LSLEAFSPPYL
Sbjct: 33 QFELQNPTKTPWMYHSTAVLLRDGRVLVAGSNPHEYYNFSSVLFPTELSLEAFSPPYLEP 212
Query: 460 EFDLVRPTIWHVTNKI-------LGYRVFYYVTFTVAKFASASEVSVRLLAPSFTTHSFG 512
++ VRP I T + G R+ + VT A A+ VS+ +LAP F THSF
Sbjct: 213 QYTNVRPRIVSPTPQAQTKLKYGQGLRIRFQVT---ALLAAPDSVSMTMLAPPFNTHSFS 383
Query: 513 MNQRMVVLKLIGVTMVNLDIYYATVVGPSTQEIAPPGYYLLFLVHAGVPSSGEWVQLM 570
MNQR++VL+ G+ V I+ V P + +APPG+YL+F+VH G+PS G WVQ++
Sbjct: 384 MNQRLLVLEPNGLKDVGESIHEVEVTAPGSATLAPPGFYLVFVVHQGIPSEGIWVQIL 557
>BP066498
Length = 512
Score = 85.5 bits (210), Expect = 2e-17
Identities = 46/117 (39%), Positives = 65/117 (55%), Gaps = 3/117 (2%)
Frame = -1
Query: 456 YLSLEFDLVRPTIWH-VTNKILGYRVFYYVTFTVAKFA--SASEVSVRLLAPSFTTHSFG 512
YL ++FD+ P I + K L Y F+ F V A + +++ V + P FTTH F
Sbjct: 512 YLDVDFDIYXPMINEDESTKELRYGQFFETVFAVQDGAELTMNDIKVTMYFPPFTTHGFS 333
Query: 513 MNQRMVVLKLIGVTMVNLDIYYATVVGPSTQEIAPPGYYLLFLVHAGVPSSGEWVQL 569
M QR++VLKL + + + +Y + P IAPPGYYLLF+ H GVP G WV +
Sbjct: 332 MGQRLLVLKLDKLMVDSPGLYRVRIAAPPNSVIAPPGYYLLFVNHRGVPGKGMWVHI 162
>AV415391
Length = 138
Score = 64.3 bits (155), Expect = 5e-11
Identities = 25/40 (62%), Positives = 34/40 (84%)
Frame = +2
Query: 340 PSWIIENMPMARVMGDMLILPNGDVIIINGAGSGTAGWEN 379
P W++E MP+ RVM DM++LP G+V+I+NGA +GTAGWEN
Sbjct: 8 PGWVMETMPIPRVMPDMILLPTGNVMILNGAANGTAGWEN 127
>BP042912
Length = 223
Score = 57.8 bits (138), Expect = 5e-09
Identities = 25/30 (83%), Positives = 28/30 (93%)
Frame = -3
Query: 541 STQEIAPPGYYLLFLVHAGVPSSGEWVQLM 570
STQEIAP GYYLLF+VHAGVPS+G WVQ+M
Sbjct: 221 STQEIAPAGYYLLFVVHAGVPSTGSWVQVM 132
>AV417355
Length = 380
Score = 32.7 bits (73), Expect = 0.16
Identities = 22/71 (30%), Positives = 35/71 (48%), Gaps = 2/71 (2%)
Frame = +2
Query: 202 NYEFIPKTTTSSSSSSSSSIHLSFLQETNDPSENNLYPFVHLLPNGNL--FIFANTRSIL 259
N + TTT++SS S SS F + DP+ P +LP NL F FA ++
Sbjct: 149 NSSYTTTTTTTTSSGSGSSSISKFSVRSGDPAAG---PGGQILPTSNLRIFTFAELKAAT 319
Query: 260 FDYKQNVVVKE 270
+++ + V+ E
Sbjct: 320 RNFRADTVLGE 352
>TC8557 similar to UP|Q9LK52 (Q9LK52) Dbj|BAA90629.1, complete
Length = 1064
Score = 31.2 bits (69), Expect = 0.47
Identities = 20/53 (37%), Positives = 31/53 (57%)
Frame = -2
Query: 208 KTTTSSSSSSSSSIHLSFLQETNDPSENNLYPFVHLLPNGNLFIFANTRSILF 260
K + SSSSSSSSS+ S L T++ S + L + + +LF+ + R +LF
Sbjct: 559 KASVSSSSSSSSSLSSSLLTSTSESSASTLR--ATIWSSTSLFVPSKNRWLLF 407
>AW719867
Length = 496
Score = 30.0 bits (66), Expect = 1.1
Identities = 14/25 (56%), Positives = 18/25 (72%)
Frame = +1
Query: 12 ILSSLIPHFHILIVSSSSSSPSPSP 36
+LSS P FH+ + SSS+SPS SP
Sbjct: 19 LLSSPFPFFHLPPIRSSSASPSNSP 93
>AV423824
Length = 181
Score = 30.0 bits (66), Expect = 1.1
Identities = 17/35 (48%), Positives = 23/35 (65%)
Frame = +3
Query: 12 ILSSLIPHFHILIVSSSSSSPSPSPLPSLLSSPSS 46
+LSSL PHF SSS S+PSP P + ++PS+
Sbjct: 33 MLSSLFPHFASSPCSSSCSAPSP-PQVTTTTTPSA 134
>TC18517 weakly similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (4%)
Length = 493
Score = 29.6 bits (65), Expect = 1.4
Identities = 18/27 (66%), Positives = 19/27 (69%), Gaps = 3/27 (11%)
Frame = +2
Query: 26 SSSSSSPSPS---PLPSLLSSPSSNQG 49
SSSSSS SPS P PS SSPSS+ G
Sbjct: 281 SSSSSSSSPSSGTPSPSSSSSPSSSPG 361
>TC12755 similar to UP|Q9C587 (Q9C587) Replication factor C large
subunit-like protein, partial (3%)
Length = 477
Score = 29.6 bits (65), Expect = 1.4
Identities = 20/71 (28%), Positives = 35/71 (49%)
Frame = -1
Query: 205 FIPKTTTSSSSSSSSSIHLSFLQETNDPSENNLYPFVHLLPNGNLFIFANTRSILFDYKQ 264
F+P T T SSS SSSS F + + + + +PF L G+ ++ ++ +
Sbjct: 399 FLPATKTQSSSPSSSSAL*IFFKGRDSGNSSQSFPF--FLLAGSSWVSFSSFGFCLSLLK 226
Query: 265 NVVVKEFPEIP 275
++V PE+P
Sbjct: 225 YLLVIFLPELP 193
>TC15474 UP|Q7Y1B9 (Q7Y1B9) Ammonium transporter, complete
Length = 1950
Score = 29.6 bits (65), Expect = 1.4
Identities = 13/20 (65%), Positives = 16/20 (80%)
Frame = +3
Query: 26 SSSSSSPSPSPLPSLLSSPS 45
SSS+S PSPSPLP ++PS
Sbjct: 456 SSSTSGPSPSPLPESPAAPS 515
>TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, partial (22%)
Length = 505
Score = 29.3 bits (64), Expect = 1.8
Identities = 18/40 (45%), Positives = 21/40 (52%)
Frame = +3
Query: 23 LIVSSSSSSPSPSPLPSLLSSPSSNQGEWELIQPTIGISA 62
L S S+SSP+ PLPSL S S+ G PT SA
Sbjct: 141 LTTSFSTSSPASHPLPSLPSPSSAAVGPTSSTPPTSPTSA 260
>AV772566
Length = 518
Score = 29.3 bits (64), Expect = 1.8
Identities = 14/20 (70%), Positives = 15/20 (75%)
Frame = +3
Query: 26 SSSSSSPSPSPLPSLLSSPS 45
SS S+SPSPSP PS SPS
Sbjct: 333 SSKSNSPSPSPSPSPSPSPS 392
>TC17490 similar to GB|AAO64817.1|29028876|BT005882 At3g04570 {Arabidopsis
thaliana;}, partial (9%)
Length = 604
Score = 29.3 bits (64), Expect = 1.8
Identities = 17/42 (40%), Positives = 23/42 (54%), Gaps = 5/42 (11%)
Frame = -2
Query: 19 HFHILIVS-----SSSSSPSPSPLPSLLSSPSSNQGEWELIQ 55
H H+++ S SS SPS S +P L SS Q W++IQ
Sbjct: 336 HIHMIMRSINSGRCSSMSPSKSIMPHLSSSSIGQQIGWQIIQ 211
>TC9870 weakly similar to UP|NO16_MEDTR (P93328) Early nodulin 16 precursor
(N-16), partial (31%)
Length = 581
Score = 28.9 bits (63), Expect = 2.4
Identities = 17/29 (58%), Positives = 19/29 (64%), Gaps = 4/29 (13%)
Frame = +1
Query: 21 HILIVSS----SSSSPSPSPLPSLLSSPS 45
H I+SS SS SPSPSP PS SSP+
Sbjct: 220 HTEILSSNSDPSSLSPSPSPAPSSSSSPT 306
>TC10758
Length = 532
Score = 28.9 bits (63), Expect = 2.4
Identities = 13/38 (34%), Positives = 16/38 (41%)
Frame = +2
Query: 19 HFHILIVSSSSSSPSPSPLPSLLSSPSSNQGEWELIQP 56
HFH S SPSP + + S+PS W P
Sbjct: 128 HFHQPTPSHPQPSPSPQKISARSSNPSPRTNSWSFSNP 241
>BP070882
Length = 496
Score = 28.9 bits (63), Expect = 2.4
Identities = 16/37 (43%), Positives = 21/37 (56%)
Frame = +2
Query: 15 SLIPHFHILIVSSSSSSPSPSPLPSLLSSPSSNQGEW 51
S +P ++ SS SSSP + PS SSPSS+ W
Sbjct: 59 SYLPFSSHILHSSLSSSPLFNDTPSSFSSPSSSNPWW 169
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,181,504
Number of Sequences: 28460
Number of extensions: 190326
Number of successful extensions: 2580
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 1982
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2422
length of query: 570
length of database: 4,897,600
effective HSP length: 95
effective length of query: 475
effective length of database: 2,193,900
effective search space: 1042102500
effective search space used: 1042102500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148345.6


