

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.4 - phase: 0
(426 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
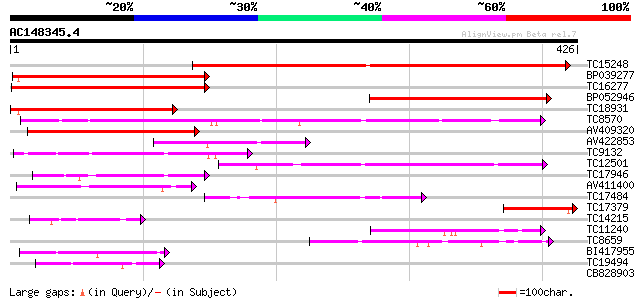
Score E
Sequences producing significant alignments: (bits) Value
TC15248 weakly similar to UP|Q8GT21 (Q8GT21) Benzoyl coenzyme A:... 292 8e-80
BP039277 217 3e-57
TC16277 weakly similar to UP|Q43583 (Q43583) Hsr201 protein, par... 184 2e-47
BP052946 182 1e-46
TC18931 similar to UP|Q8GT20 (Q8GT20) Benzoyl coenzyme A: benzyl... 180 3e-46
TC8570 similar to UP|Q9FLM5 (Q9FLM5) N-hydroxycinnamoyl/benzoylt... 180 5e-46
AV409320 176 7e-45
AV422853 106 6e-24
TC9132 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transferas... 92 2e-19
TC12501 similar to UP|Q9LR83 (Q9LR83) F21B7.2, partial (55%) 87 7e-18
TC17946 similar to UP|Q9FI59 (Q9FI59) Gb|AAD29063.1, partial (6%) 80 8e-16
AV411400 73 1e-13
TC17484 weakly similar to UP|DBAT_TAXCU (Q9M6E2) 10-deacetylbacc... 62 2e-10
TC17379 homologue to UP|Q8G4D2 (Q8G4D2) Probable ribosomal-prote... 55 2e-08
TC14215 similar to UP|Q9FYM1 (Q9FYM1) F21J9.8, partial (8%) 51 3e-07
TC11240 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transfera... 45 2e-05
TC8659 similar to UP|Q8VWP8 (Q8VWP8) Acyltransferase-like protei... 45 2e-05
BI417955 44 7e-05
TC19494 weakly similar to UP|Q9SND9 (Q9SND9) Anthranilate N-hydr... 41 3e-04
CB828903 37 0.005
>TC15248 weakly similar to UP|Q8GT21 (Q8GT21) Benzoyl coenzyme A: benzyl
alcohol benzoyl transferase, partial (52%)
Length = 1085
Score = 292 bits (747), Expect = 8e-80
Identities = 146/285 (51%), Positives = 194/285 (67%), Gaps = 1/285 (0%)
Frame = +1
Query: 138 IIDHPILLIQMARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAA 197
I+ ++++ARGA++PSI PVW+RE+L ARDPPRVT NH EYEQ+ I
Sbjct: 85 IVHFMYAVVEIARGANEPSILPVWQRELLHARDPPRVTHNHREYEQLTDDTITDPILTGT 264
Query: 198 TIVHHSFFFTLTRIAEIRRLIPFHL-RQCSTFDLITACFWCCRTKALQLEPDEEVRMMCI 256
V SFFF IA IRR +P HL + +T++++T+ W CRTKALQL+P +EVRMMCI
Sbjct: 265 DFVQQSFFFGPAEIAAIRRHLPHHLDTESTTYEVLTSYIWRCRTKALQLDPSQEVRMMCI 444
Query: 257 VNARSRFRADHSPLVGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHS 316
+AR +F GYYGNCFAFPAAV TAG+LC PL + V LI+KA +++EEYMHS
Sbjct: 445 TDARGKFNPPFP--TGYYGNCFAFPAAVATAGELCEKPLEHAVRLIKKASGEMSEEYMHS 618
Query: 317 LADLMVIKERCLFTTIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIP 376
LADLMV + + LFT +RSCVV D T A + E++FGWG+AVYGG+ AG P F +P
Sbjct: 619 LADLMVTEGKPLFTVVRSCVVLDTTYAGFRELDFGWGKAVYGGLAQAGAGAFPAVNFHVP 798
Query: 377 HKNAEGEEGLILLIFLTSEVMKRFAEELDKMFGNQNQPTTIGPSF 421
+NA+GEEG+++L+ L +++M FA+ELD M Q Q + G F
Sbjct: 799 SQNAQGEEGILVLVCLPAKIMSVFAKELDDMHA*QQQKS*SGWPF 933
>BP039277
Length = 522
Score = 217 bits (552), Expect = 3e-57
Identities = 102/150 (68%), Positives = 125/150 (83%), Gaps = 2/150 (1%)
Frame = +1
Query: 3 SSP--LLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKD 60
SSP L F V+RC+PELVPPA+ TP EVKLLSDIDDQ+ LRFN+P I +YRHEPSM +KD
Sbjct: 4 SSPPSLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKD 183
Query: 61 PAKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHP 120
P + +++ALS+TLVYYYP AGR++EG KLMVDCTGEGVMFIEA DV+L EFG+ P
Sbjct: 184 PVQAIRQALSRTLVYYYPFAGRLKEGLGRKLMVDCTGEGVMFIEANADVSLVEFGETPLP 363
Query: 121 PFPCFQELIYDVPGTNQIIDHPILLIQMAR 150
PFPCF+EL+YDVPG+ Q++D P+LLIQ+ R
Sbjct: 364 PFPCFEELLYDVPGSEQVLDSPLLLIQVTR 453
>TC16277 weakly similar to UP|Q43583 (Q43583) Hsr201 protein, partial (32%)
Length = 547
Score = 184 bits (467), Expect = 2e-47
Identities = 87/150 (58%), Positives = 115/150 (76%), Gaps = 1/150 (0%)
Frame = +1
Query: 2 TSSPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDP 61
+S PL+F+VRR +PELV PA TP E K LSDIDDQ LR N+PII YR+EPSM KDP
Sbjct: 91 SSPPLVFSVRRREPELVAPAVSTPHETKHLSDIDDQAGLRANVPIIQFYRNEPSMAGKDP 270
Query: 62 AKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPP 121
+++ A+++ LV+YYPLAGR+RE A L+VDC EGVMFIEA+ D+T ++FGD L PP
Sbjct: 271 VDIIRNAVAKALVFYYPLAGRLREAAGGNLVVDCNEEGVMFIEADADITFEQFGDTLKPP 450
Query: 122 FPCFQELIYDVPGT-NQIIDHPILLIQMAR 150
FPCFQEL+++ PG+ +I+ P++LIQ+ R
Sbjct: 451 FPCFQELLHEAPGSEGVVINSPMILIQVTR 540
>BP052946
Length = 559
Score = 182 bits (461), Expect = 1e-46
Identities = 88/137 (64%), Positives = 103/137 (74%)
Frame = -1
Query: 271 VGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFT 330
VGYYGNCF +PA VTT GKLCGN GY VEL+RKAKAQ TEEYMHS+ D +V RCLFT
Sbjct: 559 VGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMXDFLVANRRCLFT 380
Query: 331 TIRSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLI 390
T+RSC+VSDLTR K E +FGWGE V GGV AG A++II KNA+GE+G +L+I
Sbjct: 379 TVRSCIVSDLTRFKLHETDFGWGEPVCGGVAQGGAGLYGGASYIIACKNAKGEDGRVLVI 200
Query: 391 FLTSEVMKRFAEELDKM 407
L E MKRFA+EL+ M
Sbjct: 199 CLPVENMKRFAKELNNM 149
>TC18931 similar to UP|Q8GT20 (Q8GT20) Benzoyl coenzyme A: benzyl alcohol
benzoyl transferase, partial (26%)
Length = 446
Score = 180 bits (457), Expect = 3e-46
Identities = 84/128 (65%), Positives = 107/128 (82%), Gaps = 2/128 (1%)
Frame = +3
Query: 1 MTSSP--LLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIE 58
M SSP L FTVRRC+PELV P+ PTPRE+KLLSDIDDQE LRF +P+I YR++PSM
Sbjct: 63 MASSPASLAFTVRRCEPELVAPSQPTPRELKLLSDIDDQEGLRFQIPVIQFYRYDPSMTG 242
Query: 59 KDPAKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDAL 118
KDP +V+++AL++TLV+YYPLAGR+ EG KLMV+CT +GV+FIEA+ DVT+ +FGDAL
Sbjct: 243 KDPVEVIRKALAKTLVFYYPLAGRLMEGPDRKLMVNCTADGVLFIEADADVTMKQFGDAL 422
Query: 119 HPPFPCFQ 126
PPFPC++
Sbjct: 423 QPPFPCWE 446
>TC8570 similar to UP|Q9FLM5 (Q9FLM5)
N-hydroxycinnamoyl/benzoyltransferase-like protein,
partial (91%)
Length = 1551
Score = 180 bits (456), Expect = 5e-46
Identities = 127/430 (29%), Positives = 211/430 (48%), Gaps = 36/430 (8%)
Frame = +1
Query: 9 TVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRA 68
+V+ +P LV PA T + + LS++D + + ++ ++ EK +V+K A
Sbjct: 91 SVKLSEPALVLPAEETKKGMYFLSNLDQN--IAVIIRTVYCFKTAEKGNEK-AGEVVKSA 261
Query: 69 LSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQEL 128
L + LV+YYPLAGR+ KL+V+CTGEG +F+EAE + +L+E GD P +L
Sbjct: 262 LKKVLVHYYPLAGRLSISPEGKLIVECTGEGALFVEAEANCSLEEIGDITKPDPGTLGKL 441
Query: 129 IYDVPGTNQIIDHPILLIQMAR--------------------GAHQ-------------P 155
+YD+PG I+ P L+ Q+ + GA +
Sbjct: 442 VYDIPGAKHILQMPPLVAQVTKFKCGGFSLGLCMNHCMFDGIGAMEFVNSWGEAARALPL 621
Query: 156 SIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAATIVHHSFFFTLTRIAEIR 215
SI P+ R IL AR+PP++ H E+ I + N +++ SF F ++ +++
Sbjct: 622 SIPPILDRSILKARNPPKIEHLHQEFADIEDKS-NTNTLYEDEMLYRSFCFDPEKLKQLK 798
Query: 216 R--LIPFHLRQCSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPL-VG 272
+ L C+TF++++A W RTKAL+L P++E +++ V+ R F PL G
Sbjct: 799 MKAMEDGALESCTTFEVLSAFVWIARTKALKLLPEQETKLLFAVDGRKNFT---PPLPKG 969
Query: 273 YYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLFTTI 332
Y+GN +V AG+L L + V LI+ A VT+ YM S D + R +
Sbjct: 970 YFGNGIVLTNSVCQAGELSEKKLSHAVRLIQDAVKMVTDSYMRSAIDYFEV-TRARPSLA 1146
Query: 333 RSCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLIFL 392
+ +++ +R + +FGWGE V G V+ LP I+ + + + +L+ L
Sbjct: 1147CTLLITTWSRLGFHTTDFGWGEPVLSGPVS-----LPEKEVILFLSHGQERRNINVLLGL 1311
Query: 393 TSEVMKRFAE 402
+ VMK F +
Sbjct: 1312PASVMKIFQD 1341
>AV409320
Length = 392
Score = 176 bits (446), Expect = 7e-45
Identities = 80/129 (62%), Positives = 104/129 (80%)
Frame = +3
Query: 14 QPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTL 73
+PELV PA TPREVKLLSDIDDQ+ LRF +P+ YR+ PSM KDP +++AL++TL
Sbjct: 6 EPELVGPAEATPREVKLLSDIDDQDGLRFQIPVAQFYRYNPSMAGKDPVDAIRKALAKTL 185
Query: 74 VYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELIYDVP 133
V+YYP AGR+REG KLMVDCT EGV+FIEA+ DVTL+EFGD L PFPC +EL+Y+VP
Sbjct: 186 VFYYPFAGRLREGPGRKLMVDCTAEGVLFIEADADVTLNEFGDNLQTPFPCMEELLYEVP 365
Query: 134 GTNQIIDHP 142
G++++++ P
Sbjct: 366 GSDEMLNTP 392
>AV422853
Length = 456
Score = 106 bits (265), Expect = 6e-24
Identities = 61/153 (39%), Positives = 81/153 (52%), Gaps = 35/153 (22%)
Frame = +1
Query: 109 VTLDEFGDALHPPFPCFQELIYDVPGTNQIIDHPILLIQ--------------------- 147
+TL++FGD L PFPC EL+Y+VPG+ ++++ P+LLIQ
Sbjct: 1 ITLNQFGDNLQTPFPCMDELLYEVPGSEEMLNTPLLLIQVTRLKCGGFIFAIRLNHTMCD 180
Query: 148 -------------MARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEED 194
MARG QPSI PVW REIL ARD PRVTC H EY++ + KE
Sbjct: 181 AVGLVQFLSAIGEMARGMPQPSILPVWCREILSARDSPRVTCTHPEYDEQVP--YPKETT 354
Query: 195 VAA-TIVHHSFFFTLTRIAEIRRLIPFHLRQCS 226
+ +VH SFFF +A IR +P H +C+
Sbjct: 355 IPQDDMVHESFFFGPNELATIRSFLPSHQLRCT 453
>TC9132 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transferase, partial
(53%)
Length = 878
Score = 92.0 bits (227), Expect = 2e-19
Identities = 69/212 (32%), Positives = 103/212 (48%), Gaps = 33/212 (15%)
Frame = +1
Query: 4 SPLLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAK 63
S ++ TVR + +V PA P+ S++D F+ P ++ YR + D AK
Sbjct: 142 SEMIITVR--ESTMVRPAEEVPQRTVWNSNVD-LVVPNFHTPSVYFYRANGASNFFD-AK 309
Query: 64 VLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFP 123
VLK ALS+ LV +YP+AGR+R ++ +DC G+GV+F+EA+T +D+FGD P
Sbjct: 310 VLKEALSKVLVPFYPMAGRLRRDDDGRVEIDCDGQGVLFVEADTGAVIDDFGD--FAPTL 483
Query: 124 CFQELIYDVPGTNQIIDHPILLIQM---------------------ARGAH--------- 153
++LI V + I +P+L++Q+ A G H
Sbjct: 484 QQRQLIPGVDYSRGIESYPLLVLQVTHFKCGGVSLGVGMQHHVADGASGLHFINTWSDVA 663
Query: 154 ---QPSIQPVWRREILMARDPPRVTCNHHEYE 182
SI P R +L ARDPPR H EY+
Sbjct: 664 RGLDVSIPPFIDRTLLRARDPPRPAFEHIEYK 759
>TC12501 similar to UP|Q9LR83 (Q9LR83) F21B7.2, partial (55%)
Length = 1062
Score = 86.7 bits (213), Expect = 7e-18
Identities = 70/257 (27%), Positives = 114/257 (44%), Gaps = 10/257 (3%)
Frame = +3
Query: 158 QPVWRREILMARDPPRVTCNHHEYEQI-----LSSNINKEEDVAATI-VHHSFFFTLTRI 211
+P W RE+ R PP V H E+ +I L+ + + + V + F L +
Sbjct: 186 KPCWDREMFKPRHPPMVKFPHMEFMRIEEGSNLTITLWQTKPVQKCYRIQREFQNYLKTL 365
Query: 212 AEIRRLIPFHLRQCSTFDLITACFWCCRTKALQLEP-DEEVRMMCIVNARSRFRADHSPL 270
A+ P C+TFD + A W KAL + P D +R+ VNAR + R + PL
Sbjct: 366 AQ-----PSDAAGCTTFDAMAAHIWRSWVKALDVRPLDYTLRLTFSVNARPKLR--NPPL 524
Query: 271 -VGYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKERCLF 329
G+YGN ++ +L L L+R+A+ V+EEY+ S D + +
Sbjct: 525 REGFYGNVVCVACTTSSVSELVHGQLPETTRLVREARQSVSEEYLRSTVDYVDVDRPKQL 704
Query: 330 TTIRSCVVSDLTR-AKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEEG-LI 387
++ TR + Y +FGWG+ +Y G ++ P P+ +P A G +I
Sbjct: 705 EFGGKLTITQWTRFSMYKSADFGWGKPLYAGPIDLT--PTPQVCVFLPEGEAADCSGSMI 878
Query: 388 LLIFLTSEVMKRFAEEL 404
+ I L ++F + L
Sbjct: 879 VCICLPESAAQKFTQAL 929
>TC17946 similar to UP|Q9FI59 (Q9FI59) Gb|AAD29063.1, partial (6%)
Length = 699
Score = 79.7 bits (195), Expect = 8e-16
Identities = 48/135 (35%), Positives = 78/135 (57%), Gaps = 2/135 (1%)
Frame = +3
Query: 18 VPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYR--HEPSMIEKDPAKVLKRALSQTLVY 75
VPP PTP LSD+D R + PI++IY+ H+ IE+ +K +LS+ LV+
Sbjct: 144 VPPNQPTPH*HLWLSDMDQVSRQR-HTPIVYIYKAKHDQYCIER-----MKDSLSKILVH 305
Query: 76 YYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHPPFPCFQELIYDVPGT 135
YYP+AGR ++ ++C +GV+ +EAET+ T+ +FGD P +EL+ +
Sbjct: 306 YYPVAGRFSFTENGRMEINCNAKGVILLEAETEKTMADFGD--FSPSDSTKELVPTIDYN 479
Query: 136 NQIIDHPILLIQMAR 150
+ I + P+L +Q+ R
Sbjct: 480 HPIEELPLLAVQLTR 524
>AV411400
Length = 417
Score = 72.8 bits (177), Expect = 1e-13
Identities = 48/138 (34%), Positives = 71/138 (50%), Gaps = 3/138 (2%)
Frame = +2
Query: 6 LLFTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVL 65
+ +V R + +V PA TP LS +D LR N + ++RH P + +V+
Sbjct: 23 MAMSVIRTKRGMVKPAKETPLTTLDLSVMDRLPVLRCNARTLHVFRHGP-----EATRVI 187
Query: 66 KRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDE---FGDALHPPF 122
+ ALS LV YYPLAGR+ E L ++C+GEGV ++EA D TL F D P
Sbjct: 188 REALSLALVPYYPLAGRLIESKPRCLQIECSGEGVWYVEASADCTLHSVNFFDDVQSIP- 364
Query: 123 PCFQELIYDVPGTNQIID 140
+ +L+ D N+ +D
Sbjct: 365 --YDDLLPDHIPENEHVD 412
>TC17484 weakly similar to UP|DBAT_TAXCU (Q9M6E2) 10-deacetylbaccatin III
10-O-acetyltransferase (DBAT) , partial (11%)
Length = 577
Score = 61.6 bits (148), Expect = 2e-10
Identities = 46/173 (26%), Positives = 88/173 (50%), Gaps = 6/173 (3%)
Frame = +3
Query: 147 QMARGAHQPSIQPVWRREILMARDPPRVTCNHHEYEQILSSNINKEEDVAAT---IVHHS 203
++ARG ++P+I+P W R+ PP+ E E N+ + H +
Sbjct: 93 ELARGLNKPTIEPAWHRDFF----PPQ------EAEAEAQPNLKLLGPPPMPDYQLQHAN 242
Query: 204 FFFTLTRIAEIRRLIPFHLRQ-CSTFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSR 262
+ +I +++ Q CS F+++ A FW RT+A+ L+P+ +++++ N R+
Sbjct: 243 IDMPMDQIKQLKSSFEQVTGQTCSAFEIVAAKFWSSRTRAIDLDPNTQLKLVFFANCRNL 422
Query: 263 FRADHSPL-VGYYGNCFAFPAAVTTAGK-LCGNPLGYGVELIRKAKAQVTEEY 313
+ PL G+YGNCF FP +T + + L + V++I++AK ++ E+
Sbjct: 423 L---NPPLPKGFYGNCF-FPVTITASCEFLRDATIIEVVKVIQEAKGELPLEF 569
>TC17379 homologue to UP|Q8G4D2 (Q8G4D2) Probable ribosomal-protein-alanine
N-acetyltransferase, partial (6%)
Length = 519
Score = 55.1 bits (131), Expect = 2e-08
Identities = 27/57 (47%), Positives = 40/57 (69%), Gaps = 2/57 (3%)
Frame = +3
Query: 372 TFIIPHKNAEGEEGLILLIFLTSEVMKRFAEELDKMFGNQNQPTTIG--PSFVISTL 426
+F I KNA+GEEGL++ + L SE M+RF +ELD + N +PTT G SF++S++
Sbjct: 3 SFYIAFKNAKGEEGLVIPVCLPSEAMERFTKELDNVLKNHIEPTTRGFKSSFIVSSI 173
>TC14215 similar to UP|Q9FYM1 (Q9FYM1) F21J9.8, partial (8%)
Length = 1096
Score = 51.2 bits (121), Expect = 3e-07
Identities = 32/89 (35%), Positives = 49/89 (54%), Gaps = 2/89 (2%)
Frame = +1
Query: 16 ELVPPAAPTPREVKL--LSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTL 73
E++ P+ PTP +++ LS ID+ R +P+ Y H ++ LK +LS+ L
Sbjct: 118 EIIKPSTPTPPHLRIYPLSFIDNM-MYRNYIPVALFY-HPNESDKQSIISNLKNSLSEVL 291
Query: 74 VYYYPLAGRIREGARDKLMVDCTGEGVMF 102
YYP AGR+ RD+L ++C EGV F
Sbjct: 292 TRYYPFAGRL----RDQLSIECNDEGVPF 366
>TC11240 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transferase, partial
(31%)
Length = 528
Score = 45.4 bits (106), Expect = 2e-05
Identities = 39/138 (28%), Positives = 58/138 (41%), Gaps = 7/138 (5%)
Frame = +1
Query: 272 GYYGNCFAFPAAVTTAGKLCGNPLGYGVELIRKAKAQVTEEYMHSLADLMVIKE--RCLF 329
GY+GN + AG L P+ Y I A ++ EY+ S D + ++ R L
Sbjct: 43 GYFGNVIFTATPIAMAGDLMSKPIWYAASRIHDALLRMDNEYLRSALDFLELQPDLRALV 222
Query: 330 T---TIR--SCVVSDLTRAKYAEVNFGWGEAVYGGVVNCVAGPLPRATFIIPHKNAEGEE 384
T R + ++ R + +FGWG ++ G L +FIIP +G
Sbjct: 223 RGAHTFRCPNLGITSWVRLPIHDADFGWGRPIFMGPGGIAYEGL---SFIIPSPVNDG-- 387
Query: 385 GLILLIFLTSEVMKRFAE 402
L + I L E MK F E
Sbjct: 388 SLSVAIALQPEQMKVFQE 441
>TC8659 similar to UP|Q8VWP8 (Q8VWP8) Acyltransferase-like protein
(Fragment), partial (42%)
Length = 866
Score = 45.1 bits (105), Expect = 2e-05
Identities = 52/192 (27%), Positives = 77/192 (40%), Gaps = 9/192 (4%)
Frame = +3
Query: 226 STFDLITACFWCCRTKALQLEPDEEVRMMCIVNARSRFRADHSPLVGYYGNCFAFPAAVT 285
STF ++ W T A L+P++ V+ R R D Y+GN VT
Sbjct: 3 STFQALSTHIWRHVTHARSLKPEDYTVFTVFVDCRKR--VDPPMPEAYFGNLIQAIFTVT 176
Query: 286 TAGKLCGNPLGYGVELIRKA----KAQVTEEY---MHSLADLMVIKERCLFTTIRSCVVS 338
G L +P +G +I+KA A+ +E S + K+ I V
Sbjct: 177 AVGLLSAHPPQFGATMIQKAIEAHDAKAIDERNKDWESAPKIFQFKD----AGINCVTVG 344
Query: 339 DLTRAKYAEVNFGWG--EAVYGGVVNCVAGPLPRATFIIPHKNAEGEEGLILLIFLTSEV 396
R K +++FGWG E V G N G + ++ P K+ G + + + L E
Sbjct: 345 SSPRFKVYDIDFGWGKPEIVRSGPNNKFDGMI----YLYPGKS--GGRSIDVELTLEPEA 506
Query: 397 MKRFAEELDKMF 408
M R E DK F
Sbjct: 507 MGRL--EQDKEF 536
>BI417955
Length = 427
Score = 43.5 bits (101), Expect = 7e-05
Identities = 38/115 (33%), Positives = 54/115 (46%), Gaps = 2/115 (1%)
Frame = +3
Query: 8 FTVRRCQPELVPPAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKV--L 65
F+VR E+V P LS++D ++ I F Y+ S + V L
Sbjct: 24 FSVRVTNEEVVAALVPMQEHWLPLSNLD-LLLPPVDVGIFFCYKKPTSTSTTFASMVVSL 200
Query: 66 KRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEAETDVTLDEFGDALHP 120
K+AL+Q LV YY AG + + + + C GV F +A TDV L EF D +P
Sbjct: 201 KKALAQALVTYYAFAGELVVNSAGEPELLCNNRGVDFAQAVTDVEL-EFLDFYNP 362
>TC19494 weakly similar to UP|Q9SND9 (Q9SND9) Anthranilate
N-hydroxycinnamoyl/benzoyltransferase-like protein,
partial (4%)
Length = 420
Score = 41.2 bits (95), Expect = 3e-04
Identities = 30/99 (30%), Positives = 45/99 (45%), Gaps = 2/99 (2%)
Frame = +2
Query: 20 PAAPTPREVKLLSDIDDQECLRFNMPIIFIYRHEPSMIEKDPAKVLKRALSQTLVYYYPL 79
P TP LS D + + F+Y S + L+ +LS+ L +YP
Sbjct: 53 PYETTPTPTLSLSHCDQLKLPNHGSQL-FLYTPNDSPPLFNSVHTLRTSLSKVLTLFYPF 229
Query: 80 AGRI--REGARDKLMVDCTGEGVMFIEAETDVTLDEFGD 116
AGR+ G R +L+ C +G A +DV L++FGD
Sbjct: 230 AGRLCWTHGGRFELL--CNAKGAQLQGATSDVPLNDFGD 340
>CB828903
Length = 510
Score = 37.4 bits (85), Expect = 0.005
Identities = 29/105 (27%), Positives = 48/105 (45%), Gaps = 1/105 (0%)
Frame = +3
Query: 2 TSSPLLFTVRRCQPELVPPAAPTPREVKL-LSDIDDQECLRFNMPIIFIYRHEPSMIEKD 60
+SSP + + + ++P A T +KL +SD+ C +F S+
Sbjct: 84 SSSPTMLS----KTTVIPDQASTLGNLKLSVSDLPMLSCHYIQKGCLFTKPPSSSLPSHT 251
Query: 61 PAKVLKRALSQTLVYYYPLAGRIREGARDKLMVDCTGEGVMFIEA 105
+LK +LS+TL + PLAGR+ + + C GV F+ A
Sbjct: 252 LIPLLKSSLSKTLSNFPPLAGRLTTDENGYVHLTCNDAGVDFLHA 386
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.140 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,385,456
Number of Sequences: 28460
Number of extensions: 128141
Number of successful extensions: 909
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 888
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 896
length of query: 426
length of database: 4,897,600
effective HSP length: 93
effective length of query: 333
effective length of database: 2,250,820
effective search space: 749523060
effective search space used: 749523060
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148345.4


