

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.12 + phase: 0
(462 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
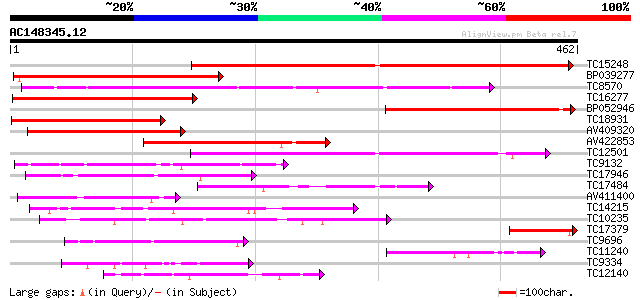
Score E
Sequences producing significant alignments: (bits) Value
TC15248 weakly similar to UP|Q8GT21 (Q8GT21) Benzoyl coenzyme A:... 317 3e-87
BP039277 258 2e-69
TC8570 similar to UP|Q9FLM5 (Q9FLM5) N-hydroxycinnamoyl/benzoylt... 235 1e-62
TC16277 weakly similar to UP|Q43583 (Q43583) Hsr201 protein, par... 202 9e-53
BP052946 184 2e-47
TC18931 similar to UP|Q8GT20 (Q8GT20) Benzoyl coenzyme A: benzyl... 181 3e-46
AV409320 180 5e-46
AV422853 174 4e-44
TC12501 similar to UP|Q9LR83 (Q9LR83) F21B7.2, partial (55%) 127 3e-30
TC9132 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transferas... 126 9e-30
TC17946 similar to UP|Q9FI59 (Q9FI59) Gb|AAD29063.1, partial (6%) 94 6e-20
TC17484 weakly similar to UP|DBAT_TAXCU (Q9M6E2) 10-deacetylbacc... 81 3e-16
AV411400 76 1e-14
TC14215 similar to UP|Q9FYM1 (Q9FYM1) F21J9.8, partial (8%) 72 1e-13
TC10235 70 1e-12
TC17379 homologue to UP|Q8G4D2 (Q8G4D2) Probable ribosomal-prote... 65 2e-11
TC9696 weakly similar to UP|Q8LAF8 (Q8LAF8) Fatty acid elongase-... 59 1e-09
TC11240 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transfera... 56 1e-08
TC9334 similar to UP|Q9MBD4 (Q9MBD4) Acyltransferase homolog, pa... 55 2e-08
TC12140 48 3e-06
>TC15248 weakly similar to UP|Q8GT21 (Q8GT21) Benzoyl coenzyme A: benzyl
alcohol benzoyl transferase, partial (52%)
Length = 1085
Score = 317 bits (812), Expect = 3e-87
Identities = 169/312 (54%), Positives = 211/312 (67%), Gaps = 1/312 (0%)
Frame = +1
Query: 149 VTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQPSIQPVWNREILMARDPPY 208
VTRLKCGGFI AL NH M DG+G+ F+ A EIARGAN+PSI PVW RE+L ARDPP
Sbjct: 13 VTRLKCGGFIFALRFNHVMTDGAGIVHFMYAVVEIARGANEPSILPVWQRELLHARDPPR 192
Query: 209 ITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIAAIRRLVPLHL-SRCTAYDLIT 267
+T NHREYEQ+ T V QSFFF A IAAIRR +P HL + T Y+++T
Sbjct: 193 VTHNHREYEQLTDDTITDPILTGTDFVQQSFFFGPAEIAAIRRHLPHHLDTESTTYEVLT 372
Query: 268 ACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLIGYYGNCIAFSAAVTTAKELC 327
+ W CRTKALQ++P ++VRMMCI +AR +F N GYYGNC AF AAV TA ELC
Sbjct: 373 SYIWRCRTKALQLDPSQEVRMMCITDARGKF---NPPFPTGYYGNCFAFPAAVATAGELC 543
Query: 328 GNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFTKGRTCMVSDWTRAKFSEVNFG 387
L +AV LI+KA +++E+YMHSLAD MV + + LFT R+C+V D T A F E++FG
Sbjct: 544 EKPLEHAVRLIKKASGEMSEEYMHSLADLMVTEGKPLFTVVRSCVVLDTTYAGFRELDFG 723
Query: 388 WGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLILPICLPPEDMKRFVKELDDMLGNQ 447
WG+AVYGG A+ G G+F F V +NA+GEEG+++ +CLP + M F KELDDM Q
Sbjct: 724 WGKAVYGGLAQAGAGAFPAVNFHVPSQNAQGEEGILVLVCLPAKIMSVFAKELDDMHA*Q 903
Query: 448 NYPTMSGPSFIL 459
+ SG F L
Sbjct: 904 QQKS*SGWPFDL 939
>BP039277
Length = 522
Score = 258 bits (658), Expect = 2e-69
Identities = 122/173 (70%), Positives = 147/173 (84%), Gaps = 2/173 (1%)
Frame = +1
Query: 4 SSP--LLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKD 61
SSP L F V+R +PELV PA+ TPHEVKLLSDIDDQ+GLRFN+P I ++RHEPSM +KD
Sbjct: 4 SSPPSLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKD 183
Query: 62 PVKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQP 121
PV+ ++ ALSRTLVYYYP AGR++EG GRKLMVDCTGEGVMFIEA AD++L +FG+ P
Sbjct: 184 PVQAIRQALSRTLVYYYPFAGRLKEGLGRKLMVDCTGEGVMFIEANADVSLVEFGETPLP 363
Query: 122 PFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLR 174
PFPCF+E+L DVPGSE ++D P+ LIQVTRLKCGGFI+AL NHTM DG+GL+
Sbjct: 364 PFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLK 522
>TC8570 similar to UP|Q9FLM5 (Q9FLM5)
N-hydroxycinnamoyl/benzoyltransferase-like protein,
partial (91%)
Length = 1551
Score = 235 bits (600), Expect = 1e-62
Identities = 140/389 (35%), Positives = 223/389 (56%), Gaps = 3/389 (0%)
Frame = +1
Query: 10 TVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKHA 69
+V+ S+P LVLPA T + LS++D + + + ++ F+ EK +V+K A
Sbjct: 91 SVKLSEPALVLPAEETKKGMYFLSNLD--QNIAVIIRTVYCFKTAEKGNEKAG-EVVKSA 261
Query: 70 LSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEI 129
L + LV+YYP AGR+ KL+V+CTGEG +F+EAEA+ +L++ GD +P ++
Sbjct: 262 LKKVLVHYYPLAGRLSISPEGKLIVECTGEGALFVEAEANCSLEEIGDITKPDPGTLGKL 441
Query: 130 LCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQ 189
+ D+PG+++I+ P + QVT+ KCGGF L L +NH M DG G +FV++W E AR A
Sbjct: 442 VYDIPGAKHILQMPPLVAQVTKFKCGGFSLGLCMNHCMFDGIGAMEFVNSWGEAAR-ALP 618
Query: 190 PSIQPVWNREILMARDPPYITCNHREYEQIL-PPNTYIKEEDTTIVVHQSFFFTSAHIAA 248
SI P+ +R IL AR+PP I H+E+ I NT ED ++++SF F +
Sbjct: 619 LSIPPILDRSILKARNPPKIEHLHQEFADIEDKSNTNTLYEDE--MLYRSFCFDPEKLKQ 792
Query: 249 I--RRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSL 306
+ + + L CT +++++A W RTKAL++ P+++ +++ V+ R F+
Sbjct: 793 LKMKAMEDGALESCTTFEVLSAFVWIARTKALKLLPEQETKLLFAVDGRKNFTPPLPK-- 966
Query: 307 IGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFT 366
GY+GN I + +V A EL +L +AV LI+ A VT+ YM S D+ + R +
Sbjct: 967 -GYFGNGIVLTNSVCQAGELSEKKLSHAVRLIQDAVKMVTDSYMRSAIDYFEV-TRARPS 1140
Query: 367 KGRTCMVSDWTRAKFSEVNFGWGEAVYGG 395
T +++ W+R F +FGWGE V G
Sbjct: 1141LACTLLITTWSRLGFHTTDFGWGEPVLSG 1227
>TC16277 weakly similar to UP|Q43583 (Q43583) Hsr201 protein, partial (32%)
Length = 547
Score = 202 bits (514), Expect = 9e-53
Identities = 98/152 (64%), Positives = 123/152 (80%), Gaps = 1/152 (0%)
Frame = +1
Query: 3 SSSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDP 62
SS PL+F+VRR +PELV PA TPHE K LSDIDDQ GLR N+P+I +R+EPSM KDP
Sbjct: 91 SSPPLVFSVRRREPELVAPAVSTPHETKHLSDIDDQAGLRANVPIIQFYRNEPSMAGKDP 270
Query: 63 VKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPP 122
V ++++A+++ LV+YYP AGR+RE AG L+VDC EGVMFIEA+ADIT +QFGD L+PP
Sbjct: 271 VDIIRNAVAKALVFYYPLAGRLREAAGGNLVVDCNEEGVMFIEADADITFEQFGDTLKPP 450
Query: 123 FPCFQEILCDVPGSE-YIIDRPIRLIQVTRLK 153
FPCFQE+L + PGSE +I+ P+ LIQVTRLK
Sbjct: 451 FPCFQELLHEAPGSEGVVINSPMILIQVTRLK 546
>BP052946
Length = 559
Score = 184 bits (468), Expect = 2e-47
Identities = 88/155 (56%), Positives = 111/155 (70%)
Frame = -1
Query: 307 IGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFT 366
+GYYGNC + A VTT +LCGN GYAVEL+RKAKAQ TE+YMHS+ DF+V RCLFT
Sbjct: 559 VGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMXDFLVANRRCLFT 380
Query: 367 KGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLILPI 426
R+C+VSD TR K E +FGWGE V GG A+GG G + G ++++ KNAKGE+G +L I
Sbjct: 379 TVRSCIVSDLTRFKLHETDFGWGEPVCGGVAQGGAGLYGGASYIIACKNAKGEDGRVLVI 200
Query: 427 CLPPEDMKRFVKELDDMLGNQNYPTMSGPSFILST 461
CLP E+MKRF KEL++M+ T SG S + T
Sbjct: 199 CLPVENMKRFAKELNNMIV*TK--TQSGSSLLFLT 101
>TC18931 similar to UP|Q8GT20 (Q8GT20) Benzoyl coenzyme A: benzyl alcohol
benzoyl transferase, partial (26%)
Length = 446
Score = 181 bits (458), Expect = 3e-46
Identities = 83/126 (65%), Positives = 107/126 (84%)
Frame = +3
Query: 2 TSSSPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKD 61
+S + L FTVRR +PELV P+ PTP E+KLLSDIDDQEGLRF +PVI +R++PSM KD
Sbjct: 69 SSPASLAFTVRRCEPELVAPSQPTPRELKLLSDIDDQEGLRFQIPVIQFYRYDPSMTGKD 248
Query: 62 PVKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQP 121
PV+V++ AL++TLV+YYP AGR+ EG RKLMV+CT +GV+FIEA+AD+T+ QFGDALQP
Sbjct: 249 PVEVIRKALAKTLVFYYPLAGRLMEGPDRKLMVNCTADGVLFIEADADVTMKQFGDALQP 428
Query: 122 PFPCFQ 127
PFPC++
Sbjct: 429 PFPCWE 446
>AV409320
Length = 392
Score = 180 bits (456), Expect = 5e-46
Identities = 83/129 (64%), Positives = 106/129 (81%)
Frame = +3
Query: 15 QPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKHALSRTL 74
+PELV PA TP EVKLLSDIDDQ+GLRF +PV +R+ PSM KDPV ++ AL++TL
Sbjct: 6 EPELVGPAEATPREVKLLSDIDDQDGLRFQIPVAQFYRYNPSMAGKDPVDAIRKALAKTL 185
Query: 75 VYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEILCDVP 134
V+YYP AGR+REG GRKLMVDCT EGV+FIEA+AD+TL++FGD LQ PFPC +E+L +VP
Sbjct: 186 VFYYPFAGRLREGPGRKLMVDCTAEGVLFIEADADVTLNEFGDNLQTPFPCMEELLYEVP 365
Query: 135 GSEYIIDRP 143
GS+ +++ P
Sbjct: 366 GSDEMLNTP 392
>AV422853
Length = 456
Score = 174 bits (440), Expect = 4e-44
Identities = 89/154 (57%), Positives = 109/154 (69%), Gaps = 2/154 (1%)
Frame = +1
Query: 110 ITLDQFGDALQPPFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGD 169
ITL+QFGD LQ PFPC E+L +VPGSE +++ P+ LIQVTRLKCGGFI A+ LNHTM D
Sbjct: 1 ITLNQFGDNLQTPFPCMDELLYEVPGSEEMLNTPLLLIQVTRLKCGGFIFAIRLNHTMCD 180
Query: 170 GSGLRQFVSAWAEIARGANQPSIQPVWNREILMARDPPYITCNHREYEQIL--PPNTYIK 227
GL QF+SA E+ARG QPSI PVW REIL ARD P +TC H EY++ + P T I
Sbjct: 181 AVGLVQFLSAIGEMARGMPQPSILPVWCREILSARDSPRVTCTHPEYDEQVPYPKETTIP 360
Query: 228 EEDTTIVVHQSFFFTSAHIAAIRRLVPLHLSRCT 261
++D +VH+SFFF +A IR +P H RCT
Sbjct: 361 QDD---MVHESFFFGPNELATIRSFLPSHQLRCT 453
>TC12501 similar to UP|Q9LR83 (Q9LR83) F21B7.2, partial (55%)
Length = 1062
Score = 127 bits (320), Expect = 3e-30
Identities = 90/302 (29%), Positives = 141/302 (45%), Gaps = 9/302 (2%)
Frame = +3
Query: 148 QVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIAR-GANQPSIQPVWNREILMARDP 206
QVT+ +CGGF L L L H + DG G QF++AWA A+ G+ +P W+RE+ R P
Sbjct: 48 QVTQFRCGGFSLGLRLCHCICDGLGAMQFLAAWAATAKSGSLVIDPKPCWDREMFKPRHP 227
Query: 207 PYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIAAIRRLV-PLHLSRCTAYDL 265
P + H E+ +I + T V + + ++ L P + CT +D
Sbjct: 228 PMVKFPHMEFMRIEEGSNLTITLWQTKPVQKCYRIQREFQNYLKTLAQPSDAAGCTTFDA 407
Query: 266 ITACYWCCRTKALQVEP-DEDVRMMCIVNARSRFSANNRSSLIGYYGNCIAFSAAVTTAK 324
+ A W KAL V P D +R+ VNAR + N G+YGN + + ++
Sbjct: 408 MAAHIWRSWVKALDVRPLDYTLRLTFSVNARPKL--RNPPLREGFYGNVVCVACTTSSVS 581
Query: 325 ELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFTKGRTCMVSDWTR-AKFSE 383
EL QL L+R+A+ V+E+Y+ S D++ + G ++ WTR + +
Sbjct: 582 ELVHGQLPETTRLVREARQSVSEEYLRSTVDYVDVDRPKQLEFGGKLTITQWTRFSMYKS 761
Query: 384 VNFGWGEAVYGGAAKGGIGSFLGGT-----FLVTHKNAKGEEGLILPICLPPEDMKRFVK 438
+FGWG+ +Y G L T FL + A +I+ ICLP ++F +
Sbjct: 762 ADFGWGKPLYAGPID------LTPTPQVCVFLPEGEAADCSGSMIVCICLPESAAQKFTQ 923
Query: 439 EL 440
L
Sbjct: 924 AL 929
>TC9132 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transferase, partial
(53%)
Length = 878
Score = 126 bits (316), Expect = 9e-30
Identities = 81/226 (35%), Positives = 122/226 (53%), Gaps = 3/226 (1%)
Frame = +1
Query: 5 SPLLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVK 64
S ++ TVR S +V PA P S++D F+ P ++ +R + D K
Sbjct: 142 SEMIITVREST--MVRPAEEVPQRTVWNSNVDLVVP-NFHTPSVYFYRANGASNFFD-AK 309
Query: 65 VLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFP 124
VLK ALS+ LV +YP AGR+R ++ +DC G+GV+F+EA+ +D FGD P
Sbjct: 310 VLKEALSKVLVPFYPMAGRLRRDDDGRVEIDCDGQGVLFVEADTGAVIDDFGDFA----P 477
Query: 125 CFQEILCDVPGSEY---IIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWA 181
Q+ +PG +Y I P+ ++QVT KCGG L + + H + DG+ F++ W+
Sbjct: 478 TLQQRQL-IPGVDYSRGIESYPLLVLQVTHFKCGGVSLGVGMQHHVADGASGLHFINTWS 654
Query: 182 EIARGANQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIK 227
++ARG + SI P +R +L ARDPP H EY+ PP + K
Sbjct: 655 DVARGLD-VSIPPFIDRTLLRARDPPRPAFEHIEYK---PPPSMAK 780
>TC17946 similar to UP|Q9FI59 (Q9FI59) Gb|AAD29063.1, partial (6%)
Length = 699
Score = 93.6 bits (231), Expect = 6e-20
Identities = 59/191 (30%), Positives = 105/191 (54%), Gaps = 3/191 (1%)
Frame = +3
Query: 14 SQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKHALSRT 73
S V P PTPH LSD+D R + P++++++ + ++ ++ +K +LS+
Sbjct: 129 SASHTVPPNQPTPH*HLWLSDMDQVSRQR-HTPIVYIYK---AKHDQYCIERMKDSLSKI 296
Query: 74 LVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEILCDV 133
LV+YYP AGR ++ ++C +GV+ +EAE + T+ FGD P +E++ +
Sbjct: 297 LVHYYPVAGRFSFTENGRMEINCNAKGVILLEAETEKTMADFGDF--SPSDSTKELVPTI 470
Query: 134 PGSEYIIDRPIRLIQVTRLKC--GGFILALSLNHTMGDGSGLRQFVSAWAEIARGAN-QP 190
+ I + P+ +Q+TR GG ++ HT+ DG G +F ++WA+IARG +P
Sbjct: 471 DYNHPIEELPLLAVQLTRFNGGEGGLADGVAWVHTLSDGLGCTRFFNSWAKIARGDTLEP 650
Query: 191 SIQPVWNREIL 201
+P +R +L
Sbjct: 651 HEKPSLDRTLL 683
>TC17484 weakly similar to UP|DBAT_TAXCU (Q9M6E2) 10-deacetylbaccatin III
10-O-acetyltransferase (DBAT) , partial (11%)
Length = 577
Score = 81.3 bits (199), Expect = 3e-16
Identities = 54/206 (26%), Positives = 100/206 (48%), Gaps = 14/206 (6%)
Frame = +3
Query: 154 CGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQPSIQPVWNREILMARD-------- 205
CGGF++ L H++ DG G QF++A E+ARG N+P+I+P W+R+ ++
Sbjct: 9 CGGFVIGLMFCHSICDGLGAAQFLNAVGELARGLNKPTIEPAWHRDFFPPQEAEAEAQPN 188
Query: 206 -----PPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIAAIRRLVPLHLSRC 260
PP + ++ I P IK+ + SF + C
Sbjct: 189 LKLLGPPPMPDYQLQHANIDMPMDQIKQ------LKSSFEQVTG-------------QTC 311
Query: 261 TAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLIGYYGNCIAFSAAV 320
+A++++ A +W RT+A+ ++P+ ++++ N R+ N G+YGNC F +
Sbjct: 312 SAFEIVAAKFWSSRTRAIDLDPNTQLKLVFFANCRNLL---NPPLPKGFYGNCF-FPVTI 479
Query: 321 TTAKE-LCGNQLGYAVELIRKAKAQV 345
T + E L + V++I++AK ++
Sbjct: 480 TASCEFLRDATIIEVVKVIQEAKGEL 557
>AV411400
Length = 417
Score = 76.3 bits (186), Expect = 1e-14
Identities = 51/137 (37%), Positives = 75/137 (54%), Gaps = 4/137 (2%)
Frame = +2
Query: 7 LLFTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVL 66
+ +V R++ +V PA TP LS +D LR N + VFRH P + +V+
Sbjct: 23 MAMSVIRTKRGMVKPAKETPLTTLDLSVMDRLPVLRCNARTLHVFRHGP-----EATRVI 187
Query: 67 KHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQ---FGDALQPPF 123
+ ALS LV YYP AGR+ E R L ++C+GEGV ++EA AD TL F D P
Sbjct: 188 REALSLALVPYYPLAGRLIESKPRCLQIECSGEGVWYVEASADCTLHSVNFFDDVQSIP- 364
Query: 124 PCFQEILCD-VPGSEYI 139
+ ++L D +P +E++
Sbjct: 365 --YDDLLPDHIPENEHV 409
>TC14215 similar to UP|Q9FYM1 (Q9FYM1) F21J9.8, partial (8%)
Length = 1096
Score = 72.4 bits (176), Expect = 1e-13
Identities = 71/278 (25%), Positives = 120/278 (42%), Gaps = 10/278 (3%)
Frame = +1
Query: 17 ELVLPAAPTPHEVKL--LSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKHALSRTL 74
E++ P+ PTP +++ LS ID+ R +PV +F H ++ + LK++LS L
Sbjct: 118 EIIKPSTPTPPHLRIYPLSFIDNMM-YRNYIPVA-LFYHPNESDKQSIISNLKNSLSEVL 291
Query: 75 VYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEILCD-- 132
YYP AGR+R+ +L ++C EGV F E + LQ P +L
Sbjct: 292 TRYYPFAGRLRD----QLSIECNDEGVPFRVTEIK---GEQSTILQNPNETLLRLLFPDK 450
Query: 133 VPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQPSI 192
+P D I +Q+ CGG + + H MGD + FV+ WA + + +
Sbjct: 451 LPWKVTEYDESIAAVQINFFSCGGLAITACVCHKMGDATTTLNFVNDWAAMTKEKGEALT 630
Query: 193 Q---PVWNR--EILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIA 247
Q P+ N + +D P P + + + V + F F + I
Sbjct: 631 QLSLPLLNGGVSVFPHKDMPCF------------PEIVFGKGNKSNAVCRRFVFPPSKIN 774
Query: 248 AIRRLVPLH-LSRCTAYDLITACYWCCRTKALQVEPDE 284
+++ +V H + T ++ITA + AL + D+
Sbjct: 775 SLKEMVTSHGVQNPTRVEVITAWIYMRAVHALGLTFDK 888
>TC10235
Length = 1116
Score = 69.7 bits (169), Expect = 1e-12
Identities = 73/311 (23%), Positives = 131/311 (41%), Gaps = 24/311 (7%)
Frame = +1
Query: 25 TPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKHALSRTLVYYYPGAGRI 84
TP ++ LL+ Q+GL F+ P K + LKH LS L ++ P GR+
Sbjct: 163 TPWDLLLLNLESIQQGLLFHKP-----------KTDHQIHHLKHTLSTILTFFPPFTGRL 309
Query: 85 --------REGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPF-PCFQEILCDVPG 135
+ C G +F+ A A+ T D ++P + P + G
Sbjct: 310 IITNHHDNNNNTTVTCHITCNNAGALFVHAAAEKT--SIADIIRPGYVPSIVHSFFPLNG 483
Query: 136 SEYI--IDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQPSIQ 193
+ + +P+ +QVT L G FI ++NH++ DG QF+++W E++RG + ++
Sbjct: 484 VKNYQGMSQPLLAVQVTELLDGVFI-GFTMNHSVADGKAFWQFINSWGELSRGNSDKLMK 660
Query: 194 -PVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQ-----SFFFTSAHIA 247
P R DPP + + E+ K++D + V Q F F+ +A
Sbjct: 661 TPPLERWFPNGIDPPIHFPSFTKEEE-------KKKQDNSDPVPQLPPLCIFHFSKEKLA 819
Query: 248 AIRRLV------PLHLSR-CTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSA 300
++ +H +R ++ + W +A +V P+++ M I++AR R
Sbjct: 820 ELKAKANAEVDNGMHENRVISSLQALLTHIWRSVIRAQRVNPEKETDYMLIIDARQRMQP 999
Query: 301 NNRSSLIGYYG 311
+ +G G
Sbjct: 1000PLPENYLGNAG 1032
>TC17379 homologue to UP|Q8G4D2 (Q8G4D2) Probable ribosomal-protein-alanine
N-acetyltransferase, partial (6%)
Length = 519
Score = 65.1 bits (157), Expect = 2e-11
Identities = 31/57 (54%), Positives = 42/57 (73%), Gaps = 2/57 (3%)
Frame = +3
Query: 408 TFLVTHKNAKGEEGLILPICLPPEDMKRFVKELDDMLGNQNYPTMSG--PSFILSTL 462
+F + KNAKGEEGL++P+CLP E M+RF KELD++L N PT G SFI+S++
Sbjct: 3 SFYIAFKNAKGEEGLVIPVCLPSEAMERFTKELDNVLKNHIEPTTRGFKSSFIVSSI 173
>TC9696 weakly similar to UP|Q8LAF8 (Q8LAF8) Fatty acid elongase-like
protein (cer2-like), partial (23%)
Length = 881
Score = 59.3 bits (142), Expect = 1e-09
Identities = 41/152 (26%), Positives = 70/152 (45%), Gaps = 2/152 (1%)
Frame = +3
Query: 45 LPVIFVFRHEPSMKEKDPVKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFI 104
L V++ F E + + +K+ +H + + +YY GR R + + C G FI
Sbjct: 216 LRVVYFFDSEAA-QGLTTMKIKEH-MFQWFNHYYFTCGRFRRSESGRPFIKCNDCGSRFI 389
Query: 105 EAEADITLDQFGDALQPPFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLN 164
EA+ TLD++ +P ++ ++ + P L Q+T+ KCGG L LS
Sbjct: 390 EAKCKKTLDEW--LAMKDWPLYKLLVSQQVIGPELSFSPSVLFQLTQFKCGGISLGLSWA 563
Query: 165 HTMGDGSGLRQFVSAWAEIA--RGANQPSIQP 194
H +GD F+++W ++ G PS P
Sbjct: 564 HVLGDPLSASDFINSWGQVMSNMGMKTPSNSP 659
>TC11240 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transferase, partial
(31%)
Length = 528
Score = 55.8 bits (133), Expect = 1e-08
Identities = 38/136 (27%), Positives = 64/136 (46%), Gaps = 7/136 (5%)
Frame = +1
Query: 308 GYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKE--RCLF 365
GY+GN I + + A +L + YA I A ++ +Y+ S DF+ ++ R L
Sbjct: 43 GYFGNVIFTATPIAMAGDLMSKPIWYAASRIHDALLRMDNEYLRSALDFLELQPDLRALV 222
Query: 366 TKGRTCM-----VSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEE 420
T ++ W R + +FGWG ++ G GGI ++ G +F++ + +
Sbjct: 223 RGAHTFRCPNLGITSWVRLPIHDADFGWGRPIFMG--PGGI-AYEGLSFII--PSPVNDG 387
Query: 421 GLILPICLPPEDMKRF 436
L + I L PE MK F
Sbjct: 388 SLSVAIALQPEQMKVF 435
>TC9334 similar to UP|Q9MBD4 (Q9MBD4) Acyltransferase homolog, partial
(47%)
Length = 817
Score = 55.1 bits (131), Expect = 2e-08
Identities = 53/170 (31%), Positives = 83/170 (48%), Gaps = 14/170 (8%)
Frame = +2
Query: 43 FNLPVIFVFRHEPSMKEKDP-----VKVLKHALSRTLVYYYPGAGRI---REGAGRKLMV 94
F+LP + + ++ + K V+ LK L+ L ++ G+I EG R +
Sbjct: 134 FDLPYLAFYYNQKLLFYKGEGFEGLVEKLKDGLAVVLKEFHQLGGKIGKDEEGVFR-VEF 310
Query: 95 DCTGEGVMFIEAEAD------ITLDQFGDALQPPFPCFQEILCDVPGSEYIIDRPIRLIQ 148
D GV IEA AD +TL + L+ P + +L ++ G + RP+ +Q
Sbjct: 311 DDDLPGVEVIEAVADEVAVADLTLTESSSILKELIP-YSGVL-NLEG----MHRPLLAVQ 472
Query: 149 VTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQPSIQPVWNR 198
T+LK G + L+ NH + DG+ F+S+WAEI GA PS+QP R
Sbjct: 473 FTKLK-DGLAMGLAFNHAVLDGTSTWHFMSSWAEICCGAPFPSVQPFQER 619
>TC12140
Length = 542
Score = 48.1 bits (113), Expect = 3e-06
Identities = 42/187 (22%), Positives = 80/187 (42%), Gaps = 7/187 (3%)
Frame = +3
Query: 77 YYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEILCDVPGS 136
YYP AGR+ + + ++C +GV+F+ IT + Q P L G
Sbjct: 27 YYPFAGRLID----RRSIECNDQGVLFLVTR--ITGTKLSTIFQNPTETLLNPLYP-DGL 185
Query: 137 EYIIDRPIR---LIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQPSIQ 193
++ + I IQ+ CGG +++ ++H +G+ + L FV WA + R +
Sbjct: 186 QWQVMSSIESILAIQINCFACGGMAISVCMSHKIGEAATLFNFVHDWATVNREKQGELLL 365
Query: 194 PVWNREILMARDPPYITCNHREYEQ----ILPPNTYIKEEDTTIVVHQSFFFTSAHIAAI 249
P P++ + Q + P + K+ +T+V + F F ++ I ++
Sbjct: 366 P-----------SPFLDGGVSLFPQGDLPVFPEMVFAKDNSSTLVCRR-FVFPASKIKSL 509
Query: 250 RRLVPLH 256
+ +V H
Sbjct: 510 KAMVSSH 530
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.139 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,667,161
Number of Sequences: 28460
Number of extensions: 126135
Number of successful extensions: 639
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 610
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 616
length of query: 462
length of database: 4,897,600
effective HSP length: 93
effective length of query: 369
effective length of database: 2,250,820
effective search space: 830552580
effective search space used: 830552580
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148345.12


