

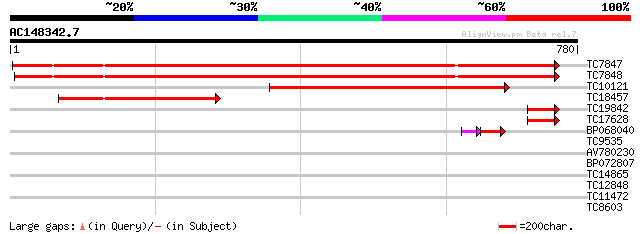
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148342.7 - phase: 0
(780 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC7847 homologue to UP|SUSY_SOYBN (P13708) Sucrose synthase (Su... 860 0.0
TC7848 homologue to UP|Q9AVR8 (Q9AVR8) Sucrose synthase isoform ... 854 0.0
TC10121 homologue to UP|SUS2_PEA (O24301) Sucrose synthase 2 (S... 456 e-129
TC18457 similar to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Suc... 195 3e-50
TC19842 similar to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Suc... 62 5e-10
TC17628 homologue to UP|SUS2_PEA (O24301) Sucrose synthase 2 (S... 61 8e-10
BP068040 44 3e-06
TC9535 38 0.007
AV780230 32 0.30
BP072807 28 7.3
TC14865 UP|Q93VA1 (Q93VA1) Selenium binding protein, complete 27 9.6
TC12848 GB|AAA37921.1|553942|MUSIGF2RA insulin-like growth facto... 27 9.6
TC11472 weakly similar to UP|Q9LYR8 (Q9LYR8) Auxin reponsive-lik... 27 9.6
TC8603 similar to UP|Q8LBJ5 (Q8LBJ5) Nucleic acid binding protei... 27 9.6
>TC7847 homologue to UP|SUSY_SOYBN (P13708) Sucrose synthase (Sucrose-UDP
glucosyltransferase) (Nodulin-100) , complete
Length = 2797
Score = 860 bits (2223), Expect = 0.0
Identities = 422/753 (56%), Positives = 556/753 (73%), Gaps = 1/753 (0%)
Frame = +1
Query: 5 HALKRTNSIADNMPDALRKSRYHMKKCFAKYLEKGRRIMKLHELMEEVERTIDDINERNY 64
+ L R +S+ + + + L +R + ++ KG+ I++ H+++ E E ++ R
Sbjct: 91 NGLTRVHSLRERLDETLATNRNEILTLLSRIEAKGKGILQHHQIIAEFEEIPEE--NRQK 264
Query: 65 ILEGNLGFILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKER 124
+ +G G +L STQEA+V PP+VA A+RP PGVWEY+RVN L VE + P ++L FKE
Sbjct: 265 LTDGAFGEVLRSTQEAIVLPPWVALAVRPRPGVWEYLRVNVHALVVEELQPAEFLHFKEE 444
Query: 125 VYDQKWANDENAFEADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIVD 184
+ D +N E DF F+ P+ TL+ SIGNG+ F+++ L+++ +++
Sbjct: 445 LVDGS-SNGNFVLELDFEPFNASFPRPTLNKSIGNGVEFLNRHLSAKLFHDKESLHPLLE 621
Query: 185 YLLKLNHHGESLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKGW 244
+L N++G++LM+ND + + LQ L A+ +L +P +T Y +FE + +E G E+GW
Sbjct: 622 FLRLHNYNGKTLMLNDRIQTPNALQHVLRKAEEYLGTLPLETPYSEFEHKFQEIGLERGW 801
Query: 245 GDNAGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPDTG 304
GD A RV E+++ L ++L+APDP LE F RIP +F VVI S HGYF Q +VLG PDTG
Sbjct: 802 GDTAERVLESIQLLLDLLEAPDPCTLETFLGRIPMVFNVVILSPHGYFAQDNVLGYPDTG 981
Query: 305 GQVVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDTKHS 364
GQVVYILDQV+ALE E++ RIKQQGL+ P+IL++TRL+PDA GT C Q E + +T+H
Sbjct: 982 GQVVYILDQVRALESEMLQRIKQQGLDIVPRILIITRLLPDAVGTTCGQRLEKVYNTEHC 1161
Query: 365 HILRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNYTDGNLVA 424
HILRVPF EKGI+ +W+SRF+++PYLE +T+D ++ ++GKPDL++GNY+DGN+VA
Sbjct: 1162HILRVPFRNEKGIVRKWISRFEVWPYLETYTEDVAHELAKELQGKPDLIVGNYSDGNIVA 1341
Query: 425 SLMARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFIITST 484
SL+A KLG+TQ TIAHALEKTKY +SD+ WK+ + KYHFSCQF AD AMN +DFIITST
Sbjct: 1342SLLAHKLGVTQCTIAHALEKTKYPESDIYWKKFEEKYHFSCQFTADLFAMNHTDFIITST 1521
Query: 485 YQEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQR 544
+QEIAGSKD GQYESH AFTLPGL RVV GI+VFDPKFNI +PGADQ+IYFPYTE +R
Sbjct: 1522FQEIAGSKDTVGQYESHTAFTLPGLYRVVHGIDVFDPKFNIVSPGADQTIYFPYTETSRR 1701
Query: 545 HSQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRN 604
+ FHP IE+LL++ V+N EHI L D+ KPIIF+MARLD VKN++GLVEWYGKN +LR
Sbjct: 1702LTSFHPEIEELLYSSVENEEHICVLKDRSKPIIFTMARLDRVKNITGLVEWYGKNAKLRE 1881
Query: 605 LVNLVIVGGFFDPSK-SKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRC 663
LVNLV+V G D K SKD EE AE+KKM+ LIE Y+L GQFRWI++Q +R RNGELYR
Sbjct: 1882LVNLVVVAG--DRRKESKDLEEKAEMKKMYSLIETYKLNGQFRWISSQMNRVRNGELYRV 2055
Query: 664 IADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDES 723
I DTKGAFVQPA+YEAFGLTV+EAM CGLPTFAT GGPAEIIV G SGFHIDP +GD +
Sbjct: 2056ICDTKGAFVQPAVYEAFGLTVVEAMTCGLPTFATCNGGPAEIIVHGKSGFHIDPYHGDRA 2235
Query: 724 SNKISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
++ + +FFEK KVDP++W+ IS GLQRI E Y
Sbjct: 2236ADLLVEFFEKVKVDPTHWDHISHGGLQRIEEKY 2334
>TC7848 homologue to UP|Q9AVR8 (Q9AVR8) Sucrose synthase isoform 3 ,
complete
Length = 2779
Score = 854 bits (2207), Expect = 0.0
Identities = 419/751 (55%), Positives = 553/751 (72%), Gaps = 1/751 (0%)
Frame = +1
Query: 7 LKRTNSIADNMPDALRKSRYHMKKCFAKYLEKGRRIMKLHELMEEVERTIDDINERNYIL 66
L R++S + + + L + + ++ + KG+ I++ ++++ E E ++ R +
Sbjct: 91 LTRSHSFRERLDETLAGQKNEILTLLSRIVAKGKGILQQNQIIAEFEEIPEE--SRQKLF 264
Query: 67 EGNLGFILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKERVY 126
G G IL STQEA+V PP+VA A+RP PGVWEY+RV+ + V+ + P +YLKFKE +
Sbjct: 265 NGAFGEILRSTQEAIVLPPFVALAVRPRPGVWEYLRVDVYSIVVDELRPAEYLKFKEELV 444
Query: 127 DQKWANDENAFEADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIVDYL 186
D +N E DF F+ P+ TL+ SIGNG+ F+++ L+++ Q ++++L
Sbjct: 445 DGS-SNGNFVLELDFEPFNASFPRPTLNKSIGNGVEFLNRHLSAKLFHGKESLQPLLEFL 621
Query: 187 LKLNHHGESLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKGWGD 246
+++G+++M+ND + + LQ L A+ +L ++ +T Y +FE + +E G E+GWGD
Sbjct: 622 RLHSYNGQTMMLNDRVQNLNSLQHVLRKAEEYLISVAPETPYSEFEHKFQEIGLERGWGD 801
Query: 247 NAGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPDTGGQ 306
A RV E ++ L ++ +APDP LE F RIP +F VVI S HGYF Q +VLG PDTGGQ
Sbjct: 802 TAERVLEMIQLLLDLFEAPDPFTLEKFLGRIPMVFNVVILSPHGYFAQDNVLGYPDTGGQ 981
Query: 307 VVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDTKHSHI 366
VVYILDQV+ALE E++ RIK+QGL+ P+IL++TRL+PDA GT C Q E + +T+H HI
Sbjct: 982 VVYILDQVRALENEMLSRIKKQGLDITPRILILTRLLPDAVGTTCGQRLEKVYNTEHCHI 1161
Query: 367 LRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNYTDGNLVASL 426
LRVPF TEKGI+ +W+SRF+++PYLER+T+D T++ +GKPDL++GNY DGN+VAS
Sbjct: 1162LRVPFRTEKGIVRKWISRFEVWPYLERYTEDVATELSKEWQGKPDLIVGNYRDGNMVASW 1341
Query: 427 MARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFIITSTYQ 486
+A KLG+TQ TIAHALEKTKY +SD+ WK+ + KYHFS QF AD AMN +DFIITST+Q
Sbjct: 1342LAHKLGVTQCTIAHALEKTKYPESDIYWKKFEEKYHFSAQFTADLFAMNHTDFIITSTFQ 1521
Query: 487 EIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRHS 546
EIAGSKD+ GQYESH AFTLPGL RVV GI+VFDPKFNI +PGAD IYFPYTE ++R +
Sbjct: 1522EIAGSKDKVGQYESHTAFTLPGLYRVVHGIDVFDPKFNIVSPGADMDIYFPYTETERRLT 1701
Query: 547 QFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLV 606
FHP IE+LLF+ V+N EHI L D+ KPIIF+MARLD VKN++GLVEWYGKN RLR LV
Sbjct: 1702HFHPDIEELLFSSVENEEHICVLKDRNKPIIFTMARLDRVKNITGLVEWYGKNARLRELV 1881
Query: 607 NLVIVGGFFDPSK-SKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCIA 665
NLV+V G D K SKD EE+AE+KKM+ LIE Y+L GQFRWI++Q DR RNGELYR I
Sbjct: 1882NLVVVAG--DRRKESKDLEEIAEMKKMYGLIETYKLNGQFRWISSQMDRIRNGELYRVIC 2055
Query: 666 DTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESSN 725
DTKGAFVQPA+YEAFGLTV+EAM CGLPTFAT GGPAEIIV G SG+HIDP +GD ++
Sbjct: 2056DTKGAFVQPAIYEAFGLTVVEAMTCGLPTFATCNGGPAEIIVHGKSGYHIDPYHGDRAAE 2235
Query: 726 KISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
+ +FFEK K DPSYW+ IS GLQRI+E Y
Sbjct: 2236TLVEFFEKSKADPSYWDKISQGGLQRIHEKY 2328
>TC10121 homologue to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Sucrose-UDP
glucosyltransferase 2) , partial (41%)
Length = 996
Score = 456 bits (1172), Expect = e-129
Identities = 214/330 (64%), Positives = 271/330 (81%)
Frame = +2
Query: 358 INDTKHSHILRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNY 417
++ T ++HILRVPF ++ G L +W+SRFD++PYLE +++D ++I ++G PD +IGNY
Sbjct: 5 VSGTDYTHILRVPFRSDNGTLHKWISRFDVWPYLETYSEDVASEITAELQGYPDFIIGNY 184
Query: 418 TDGNLVASLMARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSS 477
+DGNLVASL+A K+G+TQ TIAHALEKTKY DSD+ WK+ + KYHF+CQF AD +AMN++
Sbjct: 185 SDGNLVASLLAYKMGVTQCTIAHALEKTKYPDSDIYWKKFEDKYHFACQFTADLIAMNNA 364
Query: 478 DFIITSTYQEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFP 537
DFIITSTYQEIAG+K+ GQYESH++FTLPGL RVV GINVFDPKFNI +PGAD SIYFP
Sbjct: 365 DFIITSTYQEIAGTKNTVGQYESHSSFTLPGLYRVVHGINVFDPKFNIVSPGADMSIYFP 544
Query: 538 YTEKDQRHSQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYG 597
Y+EK +R + H +IE LL++ ++ IG L D+ KPIIFS+ARLD VKN++GLVE Y
Sbjct: 545 YSEKQKRLTALHSSIEKLLYDSEQTDDCIGSLKDRSKPIIFSIARLDRVKNITGLVESYA 724
Query: 598 KNKRLRNLVNLVIVGGFFDPSKSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRN 657
KN +LR LVNLVIV G+ D KS+DREE+AEI+KMH+L++ Y L G FRWI +QT+R RN
Sbjct: 725 KNNKLRELVNLVIVAGYIDVKKSRDREEIAEIEKMHELMKNYNLNGDFRWIVSQTNRARN 904
Query: 658 GELYRCIADTKGAFVQPALYEAFGLTVIEA 687
GELYR IADTKGAFVQPA YEAFGLTV+EA
Sbjct: 905 GELYRYIADTKGAFVQPAFYEAFGLTVVEA 994
>TC18457 similar to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Sucrose-UDP
glucosyltransferase 2) , partial (28%)
Length = 689
Score = 195 bits (495), Expect = 3e-50
Identities = 102/222 (45%), Positives = 139/222 (61%)
Frame = +3
Query: 68 GNLGFILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKERVYD 127
G G I+ S QEA+V PP+VA A+RP PGVWEYVRVN +LSVE ++ +YL FKE + D
Sbjct: 27 GPFGDIIKSAQEAIVLPPFVAIAVRPRPGVWEYVRVNVFELSVEQLSVAEYLSFKEELVD 206
Query: 128 QKWANDENAFEADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIVDYLL 187
K ND E DF +F+ + T SSSIGNG+ F+++ L+S + ++D+L
Sbjct: 207 GK-VNDNFVLELDFESFNATLTCPTRSSSIGNGVQFLNRHLSSSMFHNKDSLEPLLDFLR 383
Query: 188 KLNHHGESLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKGWGDN 247
+ G +LM+ND + + +KLQ AL A+ LS + D+ Y +FE L+ GFE+GWGD
Sbjct: 384 AHKYKGHALMLNDRIKTISKLQSALAKAEDHLSKLAPDSLYSEFEYILQGMGFERGWGDT 563
Query: 248 AGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVH 289
A RV E M L ++L APDP LE F R+P +F VVI S H
Sbjct: 564 AERVLEMMHLLLDILHAPDPSTLETFLGRVPMVFNVVILSPH 689
>TC19842 similar to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Sucrose-UDP
glucosyltransferase 2) , partial (12%)
Length = 688
Score = 61.6 bits (148), Expect = 5e-10
Identities = 26/44 (59%), Positives = 34/44 (77%)
Frame = +1
Query: 713 FHIDPLNGDESSNKISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
FHIDP + D++SN + +FF++CK DPS+WN IS GLQRI E Y
Sbjct: 1 FHIDPYHPDQASNLLXEFFQRCKEDPSHWNKISDXGLQRIYERY 132
>TC17628 homologue to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Sucrose-UDP
glucosyltransferase 2) , partial (12%)
Length = 607
Score = 60.8 bits (146), Expect = 8e-10
Identities = 26/44 (59%), Positives = 34/44 (77%)
Frame = +3
Query: 713 FHIDPLNGDESSNKISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
FHIDP + D++SN + +FF++CK DPS+WN IS GLQRI E Y
Sbjct: 3 FHIDPYHPDQASNLLVEFFQRCKEDPSHWNKISDGGLQRIYERY 134
>BP068040
Length = 431
Score = 43.5 bits (101), Expect(2) = 3e-06
Identities = 22/36 (61%), Positives = 27/36 (74%), Gaps = 2/36 (5%)
Frame = +1
Query: 648 IAAQTDRYRNGELYRCIADTKGAFVQPALYE--AFG 681
+AAQT+RYR GELYR IAD+KG F +Y+ AFG
Sbjct: 82 LAAQTNRYRKGELYRGIADSKGGFCGXPVYDMKAFG 189
Score = 38.9 bits (89), Expect = 0.003
Identities = 20/35 (57%), Positives = 24/35 (68%), Gaps = 2/35 (5%)
Frame = +2
Query: 624 EEMAEIKKMHDLIEKYQLKG--QFRWIAAQTDRYR 656
EE+AEIKKMHDLIEK+Q +G Q W+ D R
Sbjct: 8 EEVAEIKKMHDLIEKHQAQGPIQMDWLRRLIDTGR 112
Score = 25.0 bits (53), Expect(2) = 3e-06
Identities = 11/27 (40%), Positives = 16/27 (58%)
Frame = +3
Query: 622 DREEMAEIKKMHDLIEKYQLKGQFRWI 648
DR + K+ ++ +LKGQFRWI
Sbjct: 3 DRRKWQR*KRCMT*LKNTKLKGQFRWI 83
>TC9535
Length = 1083
Score = 37.7 bits (86), Expect = 0.007
Identities = 31/109 (28%), Positives = 48/109 (43%), Gaps = 3/109 (2%)
Frame = +3
Query: 622 DREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALYEAFG 681
DR A I + D + +L+ F + A GE + FV P+ E G
Sbjct: 297 DRLPEARIAFIGDGPYREELEKLFEGMPAVFTGMLGGEELSQAYASGDVFVMPSESETLG 476
Query: 682 LTVIEAMNCGLPTFATNQGGPAEII---VDGVSGFHIDPLNGDESSNKI 727
L V+EAM+ G+P GG +II DG G+ P + ++ +K+
Sbjct: 477 LVVLEAMSSGIPVVGARAGGIPDIIPEDQDGKIGYLYTPGDLEDCLSKL 623
>AV780230
Length = 583
Score = 32.3 bits (72), Expect = 0.30
Identities = 15/22 (68%), Positives = 17/22 (77%)
Frame = -1
Query: 735 KVDPSYWNVISMAGLQRINEWY 756
KVDP+ W+ IS AGLQRI E Y
Sbjct: 583 KVDPTPWDHISHAGLQRIEEKY 518
>BP072807
Length = 468
Score = 27.7 bits (60), Expect = 7.3
Identities = 13/27 (48%), Positives = 16/27 (59%)
Frame = +1
Query: 349 TKCHQEFEPINDTKHSHILRVPFHTEK 375
TK + F INDTK +I P+ TEK
Sbjct: 187 TKRYSSFCLINDTKRKNIYSCPYKTEK 267
>TC14865 UP|Q93VA1 (Q93VA1) Selenium binding protein, complete
Length = 1678
Score = 27.3 bits (59), Expect = 9.6
Identities = 27/89 (30%), Positives = 44/89 (49%), Gaps = 5/89 (5%)
Frame = +3
Query: 293 GQADVLGLPDTGGQVVYILDQVKALEEELILRIKQQGLNYKPQILVVT----RL-IPDAR 347
GQ V GL G V I + K + + + I+ Q L PQ++ ++ RL + ++
Sbjct: 1128 GQLWVGGLIQKGSPVAAIGEDGKTWQSD-VPEIQGQKLRGGPQMIQLSLDGKRLYVTNSL 1304
Query: 348 GTKCHQEFEPINDTKHSHILRVPFHTEKG 376
+ ++F P K SHIL++ TEKG
Sbjct: 1305 FSAWDKQFYPGLVEKGSHILQIDVDTEKG 1391
>TC12848 GB|AAA37921.1|553942|MUSIGF2RA insulin-like growth factor 2
receptor {Mus musculus;}, partial (23%)
Length = 664
Score = 27.3 bits (59), Expect = 9.6
Identities = 12/39 (30%), Positives = 20/39 (50%)
Frame = +3
Query: 376 GILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVI 414
G LPQW + + +F +D+ + +EGK L+I
Sbjct: 234 GFLPQWKEAWSSFRLRRKFEEDSIGFVRYWLEGKRGLII 350
>TC11472 weakly similar to UP|Q9LYR8 (Q9LYR8) Auxin reponsive-like protein,
partial (14%)
Length = 882
Score = 27.3 bits (59), Expect = 9.6
Identities = 11/32 (34%), Positives = 18/32 (55%)
Frame = -1
Query: 565 HIGYLADKRKPIIFSMARLDVVKNLSGLVEWY 596
H GYL K++ + ++RL + +L L WY
Sbjct: 408 HWGYLGLKKEAVTLELSRLSALSDLMPLGVWY 313
>TC8603 similar to UP|Q8LBJ5 (Q8LBJ5) Nucleic acid binding protein-like,
partial (88%)
Length = 1232
Score = 27.3 bits (59), Expect = 9.6
Identities = 16/38 (42%), Positives = 24/38 (63%), Gaps = 2/38 (5%)
Frame = +2
Query: 653 DRYRNGELYRCIADT--KGAFVQPALYEAFGLTVIEAM 688
D R+ +L+ CI D G+ + L +AFGLTV+EA+
Sbjct: 989 D*LRHVDLFSCI*DLCCVGSVLTFKLCDAFGLTVVEAL 1102
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.138 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,374,767
Number of Sequences: 28460
Number of extensions: 155949
Number of successful extensions: 656
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 647
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 651
length of query: 780
length of database: 4,897,600
effective HSP length: 97
effective length of query: 683
effective length of database: 2,136,980
effective search space: 1459557340
effective search space used: 1459557340
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148342.7


