

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.2 + phase: 0 /pseudo
(440 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
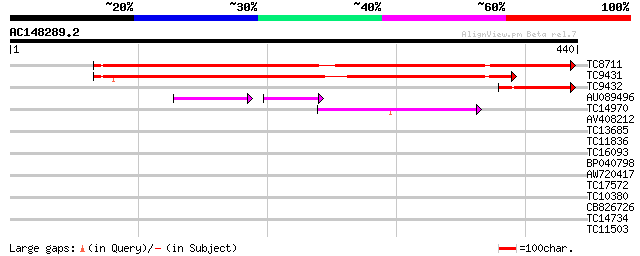
Score E
Sequences producing significant alignments: (bits) Value
TC8711 weakly similar to UP|VCLC_PEA (P13918) Vicilin precursor,... 465 e-132
TC9431 weakly similar to UP|Q84UI1 (Q84UI1) Allergen len c 3.02 ... 375 e-105
TC9432 weakly similar to UP|Q9M3X6 (Q9M3X6) Convicilin precursor... 81 3e-16
AU089496 41 2e-09
TC14970 similar to UP|O24294 (O24294) Legumin (Minor small) prec... 44 7e-05
AV408212 34 0.042
TC13685 similar to UP|Q9FJH5 (Q9FJH5) Gb|AAC98457.1 (At5g60800),... 30 0.61
TC11836 similar to GB|AAB51577.1|1755176|ATU75199 germin-like pr... 30 0.79
TC16093 weakly similar to UP|PF21_ARATH (Q04088) Possible transc... 29 1.3
BP040798 28 2.3
AW720417 28 3.0
TC17572 similar to UP|O80910 (O80910) At2g38410 protein, partial... 28 3.9
TC10380 similar to UP|Q8S8U2 (Q8S8U2) Ycf1 protein, partial (40%) 28 3.9
CB826726 27 5.1
TC14734 UP|Q9FKT4 (Q9FKT4) RNA-binding protein-like, partial (9%) 27 5.1
TC11503 similar to GB|AAO63311.1|28950775|BT005247 At3g02220 {Ar... 27 5.1
>TC8711 weakly similar to UP|VCLC_PEA (P13918) Vicilin precursor, partial
(74%)
Length = 1591
Score = 465 bits (1196), Expect = e-132
Identities = 230/374 (61%), Positives = 303/374 (80%)
Frame = +2
Query: 66 SSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHT 125
SSR Q + NPF F S+RFQT FENE+GH+R+LQRFD+RSK+FENLQNYR+LE+ +KP T
Sbjct: 449 SSRRSQ-RRNPFHFRSSRFQTHFENEHGHVRVLQRFDERSKLFENLQNYRILEFKAKPQT 625
Query: 126 LFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDL 185
L LP HNDAD I+V++SG+AILT++NPN+R+S+NLE GD + +PAG YLANRD++++L
Sbjct: 626 LVLPHHNDADSIIVILSGRAILTIVNPNDRDSYNLESGDALVIPAGATAYLANRDNDENL 805
Query: 186 RVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQE 245
RV+ L IP+NRPGQ+Q F S SE Q+S+L+GFS+NILEA+FN+ Y+EIERVL++
Sbjct: 806 RVVKLLIPINRPGQYQPFFPSSSETQESYLNGFSRNILEASFNAGYDEIERVLLQ----- 970
Query: 246 PQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPI 305
R+++R +QSQE VIVK S++QI++LS++AKSSSR+ SS+SEP NLR+ KPI
Sbjct: 971 -------REEQRGEQSQEQGVIVKASQDQIQQLSRHAKSSSRKRSSSKSEPFNLRSSKPI 1129
Query: 306 YSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFEL 365
SNKFG FEITPEKN QL+DLDIL++ A+I+EGS+ LPH++S +T+I+ V EG+GE EL
Sbjct: 1130SSNKFGKLFEITPEKNQQLRDLDILLSEAQIKEGSIFLPHYHSTSTLILVVNEGRGELEL 1309
Query: 366 VGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLG 425
V QR +Q+QR EE++++++ + QR+RARLSPGDV VIPA HP VTASSDL+LL
Sbjct: 1310VAQR---RQQQRGQEEEQEEEQPRIEAQRFRARLSPGDVIVIPASHPFAVTASSDLNLLA 1480
Query: 426 FGINAENNQRNFLA 439
FGINAENNQRNFLA
Sbjct: 1481FGINAENNQRNFLA 1522
Score = 29.6 bits (65), Expect = 1.0
Identities = 14/30 (46%), Positives = 22/30 (72%)
Frame = +2
Query: 45 IKAPCSLLMLLGIVFLASICVSSRSDQDQE 74
IKA LL+LLGI+FL S+ +++ S Q ++
Sbjct: 53 IKARLPLLLLLGILFLVSVSLATASRQRED 142
>TC9431 weakly similar to UP|Q84UI1 (Q84UI1) Allergen len c 3.02
(Fragment), partial (67%)
Length = 1328
Score = 375 bits (963), Expect = e-105
Identities = 188/330 (56%), Positives = 250/330 (74%), Gaps = 2/330 (0%)
Frame = +2
Query: 66 SSRSDQDQENPFIF--NSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKP 123
SSR Q + NPF F +S+RFQT F+NE G++R+LQRFD+RSK+FENLQNYR+ E+ +KP
Sbjct: 392 SSRRSQ-RRNPFYFRSSSSRFQTRFQNEYGYVRVLQRFDERSKLFENLQNYRIFEFKAKP 568
Query: 124 HTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNK 183
HT+ LP HNDAD I+V++SGKAI+T++NPN+R SFNLERGD + PAGT+ Y+AN DDN+
Sbjct: 569 HTVVLPHHNDADSIVVILSGKAIITLVNPNDRESFNLERGDVLVHPAGTIAYVANHDDNE 748
Query: 184 DLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENE 243
+LR+ + IPVNRPG+FQ+F S +E Q+S+L+GFS+NILEA+FN+ Y EIERVL+ E
Sbjct: 749 NLRIAKIIIPVNRPGEFQAFYPSNTEPQESYLNGFSRNILEASFNAEYNEIERVLLRGGE 928
Query: 244 QEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQK 303
Q QE +IVKVSR+ I++LS++AKSSSR+ SSE EP NLR++
Sbjct: 929 QR----------------QEQGLIVKVSRDLIQQLSRHAKSSSRKRTSSEPEPFNLRSRD 1060
Query: 304 PIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEF 363
PIYSN+FG FEI P +N Q D DI ++ EIREGS+ LPH+NSR+TVI+ V EG+GEF
Sbjct: 1061PIYSNEFGKHFEINPNRNSQPTDFDIFLSSTEIREGSIFLPHYNSRSTVILVVNEGRGEF 1240
Query: 364 ELVGQRNENQQEQREYEEDEQQQERSQQVQ 393
ELV QR QQ+QR EEDE+++E +++
Sbjct: 1241ELVAQR--KQQQQRRNEEDEEEEEEQPRIE 1324
>TC9432 weakly similar to UP|Q9M3X6 (Q9M3X6) Convicilin precursor, partial
(18%)
Length = 516
Score = 81.3 bits (199), Expect = 3e-16
Identities = 39/60 (65%), Positives = 49/60 (81%)
Frame = +1
Query: 380 EEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
EE+E+ Q R + QR+RARLSPGDV VIPAGHP+ + ASSDL+ + FGI+AENN R+FLA
Sbjct: 4 EEEEEVQPRIE-AQRFRARLSPGDVVVIPAGHPVAINASSDLNFIAFGIDAENNPRHFLA 180
>AU089496
Length = 438
Score = 40.8 bits (94), Expect(2) = 2e-09
Identities = 18/61 (29%), Positives = 36/61 (58%)
Frame = +3
Query: 128 LPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRV 187
+PQ+ D+ ++ V SG+A + + + L+RGD ++PAG+ YL N +++ L +
Sbjct: 9 VPQYLDSSLVMFVRSGEAKVAFIYKDKLAERKLKRGDVYEIPAGSAFYLVNTGESQKLHI 188
Query: 188 L 188
+
Sbjct: 189 I 191
Score = 37.4 bits (85), Expect(2) = 2e-09
Identities = 17/46 (36%), Positives = 25/46 (53%)
Frame = +1
Query: 198 GQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENE 243
G F+SF + N S LSGF ILE FN+ E+ ++ ++E
Sbjct: 226 GIFESFYIGGGANPASVLSGFEPEILETVFNATGPELRKIFTRQHE 363
>TC14970 similar to UP|O24294 (O24294) Legumin (Minor small) precursor,
partial (51%)
Length = 1566
Score = 43.5 bits (101), Expect = 7e-05
Identities = 31/132 (23%), Positives = 57/132 (42%), Gaps = 5/132 (3%)
Frame = +1
Query: 240 EENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSES----- 294
EE E+E + RG K +R++ +E + + E + +S E +
Sbjct: 691 EEEEREERGPRGRGKHWQREEEEEEEESRRTIPSRRESRGRGGCRTSNGLEENFCTLKIH 870
Query: 295 EPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIV 354
E IN ++ +Y+ + G +I P L+ L + Y + + + PH+N A I+
Sbjct: 871 ENINRPSRADLYNPRAGRISDINSLTLPILRFLGLSAEYVNLYQNGIYGPHWNINANSII 1050
Query: 355 AVEEGKGEFELV 366
V G+G +V
Sbjct: 1051YVVRGRGRVRIV 1086
>AV408212
Length = 362
Score = 34.3 bits (77), Expect = 0.042
Identities = 15/35 (42%), Positives = 24/35 (67%)
Frame = +3
Query: 368 QRNENQQEQREYEEDEQQQERSQQVQRYRARLSPG 402
Q+ + QQ+Q++ ++ +QQQ +SQQ R RA L G
Sbjct: 102 QQQQQQQQQQQQQQSQQQQPQSQQQSRDRAHLLNG 206
Score = 28.5 bits (62), Expect = 2.3
Identities = 10/23 (43%), Positives = 19/23 (82%)
Frame = +3
Query: 371 ENQQEQREYEEDEQQQERSQQVQ 393
+ QQ+Q++ ++ +QQQ++SQQ Q
Sbjct: 90 QQQQQQQQQQQQQQQQQQSQQQQ 158
>TC13685 similar to UP|Q9FJH5 (Q9FJH5) Gb|AAC98457.1 (At5g60800), partial
(37%)
Length = 585
Score = 30.4 bits (67), Expect = 0.61
Identities = 16/36 (44%), Positives = 22/36 (60%)
Frame = +1
Query: 353 IVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQER 388
+ A EEG+GE QR+ NQQ+ RE E+ Q+R
Sbjct: 379 VSAAEEGEGEGGEQRQRH*NQQQSREEN*REENQQR 486
>TC11836 similar to GB|AAB51577.1|1755176|ATU75199 germin-like protein
{Arabidopsis thaliana;} , partial (56%)
Length = 434
Score = 30.0 bits (66), Expect = 0.79
Identities = 16/42 (38%), Positives = 23/42 (54%)
Frame = +3
Query: 319 EKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGK 360
EK P L L + ++ + G L PH + RAT +V V EG+
Sbjct: 285 EKIPGLNTLGVSISRIDYAPGGLNPPHTHPRATEVVFVLEGQ 410
>TC16093 weakly similar to UP|PF21_ARATH (Q04088) Possible transcription
factor PosF21 (AtbZIP59), partial (17%)
Length = 570
Score = 29.3 bits (64), Expect = 1.3
Identities = 15/62 (24%), Positives = 30/62 (48%)
Frame = +1
Query: 330 LVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERS 389
++NYA G PH +S T++ A + + + + Q Q+ ++ +QQQ++
Sbjct: 7 MMNYASFGGGQQFYPHNHSMHTLLAAQQFQQLQIHPQKHQQHQHQFQQPIQQQQQQQQQP 186
Query: 390 QQ 391
Q
Sbjct: 187 IQ 192
>BP040798
Length = 521
Score = 28.5 bits (62), Expect = 2.3
Identities = 17/46 (36%), Positives = 27/46 (57%)
Frame = +1
Query: 87 LFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHN 132
L +NE+ H++ + KRS + +LQ + L+ SK H L +P HN
Sbjct: 304 LEDNEH*HLKQWLQKQKRSLLETSLQRVKPLQEASK-HWLQMPHHN 438
>AW720417
Length = 554
Score = 28.1 bits (61), Expect = 3.0
Identities = 17/56 (30%), Positives = 27/56 (47%), Gaps = 1/56 (1%)
Frame = +3
Query: 368 QRNENQQEQREYEEDEQQQERSQQVQ-RYRARLSPGDVYVIPAGHPIVVTASSDLS 422
Q +NQ + + + QQQ++ QQ Q R +L D V+ V + S+D S
Sbjct: 105 QHQQNQPQYHQQHQHNQQQQQQQQQQWLRRTQLGGNDTNVVEEVEKTVQSESADPS 272
>TC17572 similar to UP|O80910 (O80910) At2g38410 protein, partial (28%)
Length = 772
Score = 27.7 bits (60), Expect = 3.9
Identities = 17/54 (31%), Positives = 29/54 (53%)
Frame = -2
Query: 15 FIAINSTLSSQHSSSIASKLSEIISKINMAIKAPCSLLMLLGIVFLASICVSSR 68
F A + +L ++ I SKL ++ I++AI + LLGI + IC++ R
Sbjct: 393 FTACSISLKRSNTCCIDSKLEKLKLSISVAIVSSSVFDELLGIPYPGCICLAVR 232
>TC10380 similar to UP|Q8S8U2 (Q8S8U2) Ycf1 protein, partial (40%)
Length = 5664
Score = 27.7 bits (60), Expect = 3.9
Identities = 21/53 (39%), Positives = 25/53 (46%), Gaps = 11/53 (20%)
Frame = +3
Query: 75 NPFIFNSNRFQTLFENENGHIRLLQRF------DKRSKIFENLQ-----NYRL 116
NP NS +T+F+NEN Q F DK KI NL+ NYRL
Sbjct: 5166 NPRSINSMPRKTIFDNENKINNCCQVFAKNKDLDKEKKILMNLKLILWPNYRL 5324
>CB826726
Length = 505
Score = 27.3 bits (59), Expect = 5.1
Identities = 14/42 (33%), Positives = 22/42 (52%)
Frame = +2
Query: 395 YRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRN 436
+ + +SP V P ++V + ++SLLGF I NN N
Sbjct: 128 FLSTISPSSSRV*PRTTTVLVHSLCEVSLLGFSIAQMNNNNN 253
>TC14734 UP|Q9FKT4 (Q9FKT4) RNA-binding protein-like, partial (9%)
Length = 598
Score = 27.3 bits (59), Expect = 5.1
Identities = 15/58 (25%), Positives = 28/58 (47%)
Frame = -2
Query: 92 NGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTV 149
N HI+L +K K + Y++ H+ + LPQ + +I+++ S I T+
Sbjct: 564 NMHIKLTVWEEKIHKSIIYI*YYKMSPIHALKQIIHLPQSGNKPYIIIISSRNHIKTI 391
>TC11503 similar to GB|AAO63311.1|28950775|BT005247 At3g02220 {Arabidopsis
thaliana;}, partial (60%)
Length = 975
Score = 27.3 bits (59), Expect = 5.1
Identities = 20/73 (27%), Positives = 36/73 (48%), Gaps = 12/73 (16%)
Frame = +2
Query: 268 VKVSREQIEELSKNAKSSSRRS----------ESSESEPINLRNQK--PIYSNKFGNFFE 315
V+ ++ +EE KNA+ RRS +SS+ P + ++ K P++ N + +
Sbjct: 347 VEAEQKMLEEAIKNARERERRSLLRAMNKDKPKSSKDTPTDTKDMKVGPLFPN--ASLDD 520
Query: 316 ITPEKNPQLKDLD 328
+KN +DLD
Sbjct: 521 YARQKNRVNEDLD 559
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.133 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,103,638
Number of Sequences: 28460
Number of extensions: 72788
Number of successful extensions: 528
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 520
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 525
length of query: 440
length of database: 4,897,600
effective HSP length: 93
effective length of query: 347
effective length of database: 2,250,820
effective search space: 781034540
effective search space used: 781034540
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148289.2


