

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148176.1 + phase: 0
(700 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
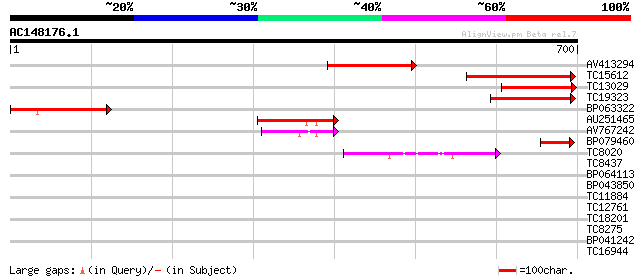
Score E
Sequences producing significant alignments: (bits) Value
AV413294 207 5e-54
TC15612 homologue to UP|Q8L693 (Q8L693) 1-deoxy-D-xylulose 5-pho... 194 4e-50
TC13029 similar to UP|Q9LFL9 (Q9LFL9) 1-D-deoxyxylulose 5-phosph... 173 1e-43
TC19323 homologue to UP|Q9XH50 (Q9XH50) 1-D-deoxyxylulose 5-phos... 157 5e-39
BP063322 147 7e-36
AU251465 100 6e-22
AV767242 82 2e-16
BP079460 57 8e-09
TC8020 homologue to UP|Q9ZQY2 (Q9ZQY2) Pyruvate dehydrogenase E1... 54 6e-08
TC8437 homologue to GB|AAB86804.1|2454184|ATU80186 pyruvate dehy... 38 0.005
BP064113 31 0.76
BP043850 30 1.00
TC11884 29 2.2
TC12761 similar to UP|Q9LIZ6 (Q9LIZ6) ESTs C28508(C61374), parti... 28 5.0
TC18201 homologue to GB|AAB86804.1|2454184|ATU80186 pyruvate deh... 28 6.5
TC8275 homologue to UP|Q94C02 (Q94C02) Myo-inositol-1-phosphate ... 28 6.5
BP041242 27 8.5
TC16944 similar to UP|Q94KL4 (Q94KL4) BEL1-like homeodomain 1, p... 27 8.5
>AV413294
Length = 333
Score = 207 bits (527), Expect = 5e-54
Identities = 99/110 (90%), Positives = 105/110 (95%)
Frame = +1
Query: 393 AEKDKDIVVVHAGITTEPSLKLFMEKFPDRIFNVGIAEQHAVTFASGLSCGGLKPFCIIP 452
AEKDKDIVVVHAG+ EPSL+LF EKFPDR F+VG+AEQHAVTFASGLSCGGLKPFCIIP
Sbjct: 4 AEKDKDIVVVHAGMLMEPSLELFREKFPDRFFDVGMAEQHAVTFASGLSCGGLKPFCIIP 183
Query: 453 SSFLQRAYDQVVHDVDQQKVPVRFVITSAGLVGSDGPLQCGAFDITFMSC 502
SSFLQRAYDQVVHDVDQQK+PVRFVITSAG+VGSDGPL CGAFDITFMSC
Sbjct: 184 SSFLQRAYDQVVHDVDQQKIPVRFVITSAGVVGSDGPLHCGAFDITFMSC 333
>TC15612 homologue to UP|Q8L693 (Q8L693) 1-deoxy-D-xylulose 5-phosphate
synthase 1 precursor, partial (19%)
Length = 703
Score = 194 bits (493), Expect = 4e-50
Identities = 95/135 (70%), Positives = 108/135 (79%)
Frame = +3
Query: 564 EGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCKHHSFLITV 623
EG+ VALLGYGS VQNCL A SL+ G+ +TVADARFCKPLD L+R L K H LITV
Sbjct: 3 EGERVALLGYGSAVQNCLAAASLVERHGLRITVADARFCKPLDRSLIRSLAKSHEVLITV 182
Query: 624 EEGSIGGFGSHVAQFIALDGLLDRRIKWRPIVLPDSYIEHASPNQQLNQAGLTGHHIAAT 683
EEGSIGGFGSHVAQF+ALDGLLD ++KWRP+VLPD YIEH SPN QL+ AGL HIAAT
Sbjct: 183 EEGSIGGFGSHVAQFMALDGLLDGKLKWRPVVLPDRYIEHGSPNDQLSSAGLAPSHIAAT 362
Query: 684 ALSLLGRTREALSFM 698
++LG+TREAL M
Sbjct: 363 VFNILGQTREALEVM 407
>TC13029 similar to UP|Q9LFL9 (Q9LFL9) 1-D-deoxyxylulose 5-phosphate
synthase-like protein, partial (13%)
Length = 504
Score = 173 bits (438), Expect = 1e-43
Identities = 83/92 (90%), Positives = 87/92 (94%)
Frame = +1
Query: 608 ELLRQLCKHHSFLITVEEGSIGGFGSHVAQFIALDGLLDRRIKWRPIVLPDSYIEHASPN 667
+LL LCKHHSFLITVEEGS+GGFGSHVAQFIALDGLLDR IKWRPIVLPD YIEHASPN
Sbjct: 1 KLLSHLCKHHSFLITVEEGSVGGFGSHVAQFIALDGLLDRGIKWRPIVLPDRYIEHASPN 180
Query: 668 QQLNQAGLTGHHIAATALSLLGRTREALSFMC 699
+QL+QAGLTGHHIAATALSLLGRTREAL FMC
Sbjct: 181 EQLDQAGLTGHHIAATALSLLGRTREALQFMC 276
>TC19323 homologue to UP|Q9XH50 (Q9XH50) 1-D-deoxyxylulose 5-phosphate
synthase, partial (15%)
Length = 530
Score = 157 bits (397), Expect = 5e-39
Identities = 76/105 (72%), Positives = 86/105 (81%)
Frame = +1
Query: 594 VTVADARFCKPLDIELLRQLCKHHSFLITVEEGSIGGFGSHVAQFIALDGLLDRRIKWRP 653
VTVADARFCKPLD L+R L K H LITVEEGSIGGFGSH QF+ALDGLLD ++KWRP
Sbjct: 4 VTVADARFCKPLDHSLIRSLAKSHEILITVEEGSIGGFGSHFVQFMALDGLLDGKLKWRP 183
Query: 654 IVLPDSYIEHASPNQQLNQAGLTGHHIAATALSLLGRTREALSFM 698
+VLPD YI+H SP QL+QAGLT HIAAT L++LG+TREAL M
Sbjct: 184 MVLPDCYIDHGSPADQLSQAGLTPSHIAATVLNMLGQTREALEVM 318
>BP063322
Length = 542
Score = 147 bits (370), Expect = 7e-36
Identities = 84/135 (62%), Positives = 98/135 (72%), Gaps = 10/135 (7%)
Frame = +3
Query: 1 MMGTVCARYPFVIPFHSQ--TLTRRLDCSISQFP----LSRITYS--SSSRILIHRVCSR 52
MMG V ARYP IP HS L+RR+D + S FP LSR S+S I RVC+R
Sbjct: 138 MMGAVPARYPLGIPLHSHGGALSRRVDFTTSHFPVHLDLSRXALCPRSASEEFIDRVCAR 317
Query: 53 PDIDDFYWE--KVPTPILDTVQNPLCLKNLSQQELKQLAAEIRLELSSILSGTQILLNPS 110
DIDDF WE +VPTPILDTV+NPLCLKNLS +ELKQLAAEIR++LSS++S TQI S
Sbjct: 318 SDIDDFDWEWDEVPTPILDTVENPLCLKNLSLKELKQLAAEIRMDLSSLMSDTQISPKAS 497
Query: 111 MAVVDLTVAIHHVFH 125
MAV +LTVA+HHVFH
Sbjct: 498 MAVAELTVAMHHVFH 542
>AU251465
Length = 363
Score = 100 bits (250), Expect = 6e-22
Identities = 54/110 (49%), Positives = 73/110 (66%), Gaps = 10/110 (9%)
Frame = +3
Query: 307 GSTLFEELGLYYIGPVDGHNIEDLISVLQEVASLDSMGPVLIHVITNENQVEEHNKKS-- 364
GS+LFEELGLYYIGPV+GHNI+DL+++L EV S + GPVLIHVIT + + + +K+
Sbjct: 9 GSSLFEELGLYYIGPVNGHNIDDLVAILNEVKSTQTTGPVLIHVITEKGRGYPYAEKAAD 188
Query: 365 -YMTDKQQDDNAGR-------LQTYGDCFVESLVAEAEKDKDIVVVHAGI 406
Y + D G+ Q+Y F E+L+AEAE DKDIV +HA +
Sbjct: 189 KYHGVTKFDPTTGKQFKAKATTQSYTTYFAEALIAEAEADKDIVAIHAAM 338
>AV767242
Length = 332
Score = 82.4 bits (202), Expect = 2e-16
Identities = 46/107 (42%), Positives = 64/107 (58%), Gaps = 11/107 (10%)
Frame = +2
Query: 311 FEELGLYYIGPVDGHNIEDLISVLQEVASLDSMGPVLIHVITNENQ----VEEHNKKSYM 366
FEELGLYYIGPVDGHN+EDL+++ ++V + + GPVLIH++T + + E + K +
Sbjct: 2 FEELGLYYIGPVDGHNVEDLVTIFEKVKATPAPGPVLIHIVTEKGKGYPPAEGADDKMHG 181
Query: 367 TDKQQDDNAGR-------LQTYGDCFVESLVAEAEKDKDIVVVHAGI 406
K D GR +Y F ESL+ EAE D IV +HA +
Sbjct: 182 VVK-FDPKTGRQFKPKPSTLSYTQYFAESLIKEAEIDNKIVAIHAAM 319
>BP079460
Length = 375
Score = 57.4 bits (137), Expect = 8e-09
Identities = 27/42 (64%), Positives = 31/42 (73%)
Frame = -1
Query: 656 LPDSYIEHASPNQQLNQAGLTGHHIAATALSLLGRTREALSF 697
LPD YIEH SP Q+++AGLT HI AT LSLL R +EAL F
Sbjct: 372 LPDRYIEHGSPQDQMDEAGLTSKHIVATVLSLLERPKEALLF 247
>TC8020 homologue to UP|Q9ZQY2 (Q9ZQY2) Pyruvate dehydrogenase E1 beta
subunit isoform 2 , partial (90%)
Length = 1643
Score = 54.3 bits (129), Expect = 6e-08
Identities = 56/209 (26%), Positives = 88/209 (41%), Gaps = 15/209 (7%)
Frame = +2
Query: 413 KLFMEKF-PDRIFNVGIAEQHAVTFASGLSCGGLKPFC-IIPSSFLQRAYDQVVHDV--- 467
K +EK+ P+R+ + I E G + GL+P + +F +A D +++
Sbjct: 362 KGLLEKYGPERVLDTPITEAGFTGIGVGAAYYGLRPVVEFMTFNFSMQAIDHIINSAAKS 541
Query: 468 -----DQQKVPVRFVITSAGLVGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVA 522
Q VP+ F + G G + + SC P + V++P + ++
Sbjct: 542 NYMSAGQINVPIVFRGPNGAAAGV-GAQHSQCYASWYGSC-PGLKVLSPYSSEDARGLLK 715
Query: 523 TAAHINDQPVCFRYPRGALVGKD-----EAILDGIPIEIGKGRILVEGKDVALLGYGSMV 577
A D PV F L G+ E + + IGK +I EGKDV + + MV
Sbjct: 716 AAIRDPD-PVVF-LENELLYGESFPVSAEVLDSSFCLPIGKAKIEREGKDVTITAFSKMV 889
Query: 578 QNCLKAYSLLANLGIEVTVADARFCKPLD 606
LKA +LA GI V + R +PLD
Sbjct: 890 GYALKAAEILAKEGISAEVINLRSIRPLD 976
>TC8437 homologue to GB|AAB86804.1|2454184|ATU80186 pyruvate dehydrogenase
E1 beta subunit {Arabidopsis thaliana;} , partial (40%)
Length = 915
Score = 38.1 bits (87), Expect = 0.005
Identities = 24/88 (27%), Positives = 41/88 (46%), Gaps = 1/88 (1%)
Frame = +1
Query: 554 IEIGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELL-RQ 612
+ + + ++ G+ V +L Y M + ++A L N G + V D R KP D+ +
Sbjct: 85 LSLEEAEMVRPGEHVTILTYSRMRYHVMQAAKTLVNKGYDPEVIDIRSLKPFDLHTIGNS 264
Query: 613 LCKHHSFLITVEEGSIGGFGSHVAQFIA 640
+ K H LI E GG G+ + I+
Sbjct: 265 VKKTHRVLIVEECMRTGGIGASLTAAIS 348
>BP064113
Length = 480
Score = 30.8 bits (68), Expect = 0.76
Identities = 16/74 (21%), Positives = 36/74 (48%)
Frame = -3
Query: 60 WEKVPTPILDTVQNPLCLKNLSQQELKQLAAEIRLELSSILSGTQILLNPSMAVVDLTVA 119
WE VP P+ DT+ N +C ++ +++ + SS+ +G Q ++ + ++ + +
Sbjct: 364 WEHVPEPVADTISNHIC--GMNHGDMQPEVEALTHYFSSLDTGGQSMVRRKLQLLFINCS 191
Query: 120 IHHVFHAPVDKILW 133
+V + K W
Sbjct: 190 YFYVNQSYFRK*PW 149
>BP043850
Length = 515
Score = 30.4 bits (67), Expect = 1.00
Identities = 16/42 (38%), Positives = 24/42 (57%)
Frame = +1
Query: 222 SNAGYLDSNLVVILNDSRHSLLPKIEDGSKTSVNALSSTLSR 263
+NAG+ N VIL + + P+I+ G TS+N L+ L R
Sbjct: 139 NNAGFACLNAAVILPANSIDIPPQIDAGDSTSINLLTRKLWR 264
>TC11884
Length = 557
Score = 29.3 bits (64), Expect = 2.2
Identities = 10/20 (50%), Positives = 13/20 (65%)
Frame = +1
Query: 60 WEKVPTPILDTVQNPLCLKN 79
WE VP P+ DT+ N +C N
Sbjct: 388 WEHVPEPVADTISNHICGMN 447
>TC12761 similar to UP|Q9LIZ6 (Q9LIZ6) ESTs C28508(C61374), partial (42%)
Length = 829
Score = 28.1 bits (61), Expect = 5.0
Identities = 14/32 (43%), Positives = 19/32 (58%)
Frame = +2
Query: 390 VAEAEKDKDIVVVHAGITTEPSLKLFMEKFPD 421
VAE E+DKD +V GI E + ++KF D
Sbjct: 311 VAEFERDKDCLVCGPGIRIELDPSITLQKFMD 406
>TC18201 homologue to GB|AAB86804.1|2454184|ATU80186 pyruvate dehydrogenase
E1 beta subunit {Arabidopsis thaliana;} , partial (25%)
Length = 636
Score = 27.7 bits (60), Expect = 6.5
Identities = 18/54 (33%), Positives = 24/54 (44%), Gaps = 1/54 (1%)
Frame = +2
Query: 587 LANLGIEVTVADARFCKPLDIELL-RQLCKHHSFLITVEEGSIGGFGSHVAQFI 639
L N G + V D R KP D+ + + K H LI E GG G+ + I
Sbjct: 5 LVNKGYDPEVIDIRSLKPFDLHTIGNSVKKTHRVLIVEECMRTGGIGASLTAAI 166
>TC8275 homologue to UP|Q94C02 (Q94C02) Myo-inositol-1-phosphate synthase
, complete
Length = 1889
Score = 27.7 bits (60), Expect = 6.5
Identities = 30/125 (24%), Positives = 51/125 (40%), Gaps = 10/125 (8%)
Frame = +2
Query: 133 WDVGDQTYAHKILTGRR---SLMKTIRK-KNGLSGFTSRFESEYDAFGAGHGCNSISAGL 188
WD+ D A + R L K +R + ++ ++ A G N++ G
Sbjct: 536 WDISDMNLADAMGRARVFDIDLQKQLRPYMESMVPLPGIYDPDFIAANQGDRANNVIKGT 715
Query: 189 G----MAVARDIKGRRE--RVVAVISNWTTMSGQVYEAMSNAGYLDSNLVVILNDSRHSL 242
+ +DIK +E +V V+ WT + + SNLVV LND+ +L
Sbjct: 716 KKEQIQQIIKDIKEFKEASKVDRVVVLWTANTERY-----------SNLVVGLNDTMENL 862
Query: 243 LPKIE 247
L ++
Sbjct: 863 LASVD 877
>BP041242
Length = 542
Score = 27.3 bits (59), Expect = 8.5
Identities = 19/56 (33%), Positives = 27/56 (47%)
Frame = +1
Query: 178 GHGCNSISAGLGMAVARDIKGRRERVVAVISNWTTMSGQVYEAMSNAGYLDSNLVV 233
GH S S G A + + R ++ I + QV EA++NAG SNL+V
Sbjct: 190 GHHHPSQSYGSAGGYAPEQRKRSDKKYTRIDDNYKSLDQVTEALANAGLESSNLIV 357
>TC16944 similar to UP|Q94KL4 (Q94KL4) BEL1-like homeodomain 1, partial
(29%)
Length = 1198
Score = 27.3 bits (59), Expect = 8.5
Identities = 12/27 (44%), Positives = 17/27 (62%)
Frame = +3
Query: 23 RLDCSISQFPLSRITYSSSSRILIHRV 49
R DC QFP S + +S++S LI R+
Sbjct: 657 REDCQNDQFPSSELGFSNTSSTLIPRI 737
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.137 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,384,040
Number of Sequences: 28460
Number of extensions: 151321
Number of successful extensions: 684
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 678
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 680
length of query: 700
length of database: 4,897,600
effective HSP length: 97
effective length of query: 603
effective length of database: 2,136,980
effective search space: 1288598940
effective search space used: 1288598940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148176.1


