

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.5 + phase: 0
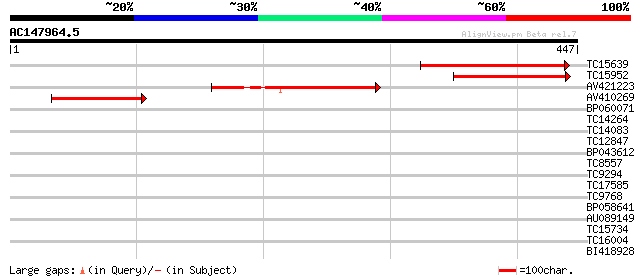
(447 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15639 weakly similar to UP|BAD05341 (BAD05341) Membrane protei... 156 7e-39
TC15952 similar to GB|AAO63445.1|28951043|BT005381 At2g25737 {Ar... 140 4e-34
AV421223 130 3e-31
AV410269 113 7e-26
BP060071 32 0.16
TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), par... 32 0.21
TC14083 similar to UP|CRTC_RICCO (P93508) Calreticulin precursor... 31 0.47
TC12847 similar to GB|AAM66074.1|21595125|AY088542 endosperm-spe... 28 2.3
BP043612 28 3.1
TC8557 similar to UP|Q9LK52 (Q9LK52) Dbj|BAA90629.1, complete 28 4.0
TC9294 homologue to UP|CAE45585 (CAE45585) Coatomer alpha subuni... 27 5.2
TC17585 homologue to UP|Q7VBB6 (Q7VBB6) Uncharacterized protein,... 27 6.8
TC9768 similar to GB|AAP68290.1|31711868|BT008851 At3g12156 {Ara... 27 6.8
BP058641 27 6.8
AU089149 27 8.9
TC15734 27 8.9
TC16004 27 8.9
BI418928 27 8.9
>TC15639 weakly similar to UP|BAD05341 (BAD05341) Membrane protein-like,
partial (25%)
Length = 556
Score = 156 bits (394), Expect = 7e-39
Identities = 75/117 (64%), Positives = 96/117 (81%)
Frame = +1
Query: 325 LGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQ 384
+GPLFLELGI PQVASAT+TF M FS+S+SVVQYY L+RFP+PYA YL LVA IAA GQ
Sbjct: 1 MGPLFLELGIAPQVASATATFGMTFSASISVVQYYLLNRFPVPYALYLTLVAAIAAYRGQ 180
Query: 385 HVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLCHH 441
++ K++ +F RAS+I+F LAFTIFVSAI LGG GI +M+ +++ EYMGF++LC +
Sbjct: 181 FIIDKLVNMFQRASLIIFTLAFTIFVSAIVLGGEGISDMIGQIQRSEYMGFEDLCKY 351
>TC15952 similar to GB|AAO63445.1|28951043|BT005381 At2g25737 {Arabidopsis
thaliana;}, partial (19%)
Length = 535
Score = 140 bits (353), Expect = 4e-34
Identities = 66/92 (71%), Positives = 80/92 (86%)
Frame = +2
Query: 351 SSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFV 410
SSMSV++YY L RFPIPYA YL LVATIAA+ GQH+VR++I +FGRAS+I+FILA TIF+
Sbjct: 2 SSMSVIEYYLLKRFPIPYAGYLTLVATIAAVVGQHIVRRLIILFGRASLIIFILAGTIFI 181
Query: 411 SAISLGGVGIGNMVEKMENEEYMGFDNLCHHG 442
SAISLGGVGI NM+ K+ N EYMGF+NLC +G
Sbjct: 182 SAISLGGVGIINMIHKIHNHEYMGFENLCKYG 277
>AV421223
Length = 414
Score = 130 bits (328), Expect = 3e-31
Identities = 67/140 (47%), Positives = 96/140 (67%), Gaps = 7/140 (5%)
Frame = +2
Query: 160 FIGTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPA------- 212
FIGTSTKA +KG++TWKKETIMKKEA A+ ES + +E +YK+LP+
Sbjct: 5 FIGTSTKAFLKGVETWKKETIMKKEA----ARRKESAGS---GAEVEYKALPSGPNNGDV 163
Query: 213 DLQDEEVPLLDNIHWKELSVLMYVWVAFLIVQILKTYSKTCSIEYWLLNSLQVPIAISVT 272
+ +++EV +L+N++WKE +L +VWV+FL +QI K TCS YW+LN LQ+PI++ VT
Sbjct: 164 ETKEQEVSILENVYWKEFGLLAFVWVSFLALQITKQNVATCSPAYWVLNFLQIPISVGVT 343
Query: 273 LFEAICLCKGTRVIASRGKE 292
+EA L G +VI+S G +
Sbjct: 344 AYEASALFSGRKVISSTGDQ 403
>AV410269
Length = 430
Score = 113 bits (282), Expect = 7e-26
Identities = 47/75 (62%), Positives = 65/75 (86%)
Frame = +1
Query: 34 SSYERVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALS 93
S +++WPE++F WR+V+ +++GF G+A G+VGGVGGGGIF+PMLTLI+GFD KS+ ALS
Sbjct: 205 SGTKKIWPELEFSWRLVLATLIGFLGSAFGTVGGVGGGGIFVPMLTLIVGFDTKSAAALS 384
Query: 94 KCMITGAAGSTVYYN 108
KCMI GA+ S+V+YN
Sbjct: 385 KCMIMGASTSSVWYN 429
>BP060071
Length = 324
Score = 32.3 bits (72), Expect = 0.16
Identities = 11/26 (42%), Positives = 19/26 (72%)
Frame = -1
Query: 416 GGVGIGNMVEKMENEEYMGFDNLCHH 441
GG GI +M+ ++ EY+GF++LC +
Sbjct: 150 GGEGISDMIGQIPRSEYLGFEDLCQY 73
>TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), partial (76%)
Length = 1277
Score = 32.0 bits (71), Expect = 0.21
Identities = 25/90 (27%), Positives = 38/90 (41%)
Frame = -3
Query: 54 IVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPKSSTALSKCMITGAAGSTVYYNLRLRH 113
+ FFGA G+GGGG F P + L+ D + A + G A T + +R
Sbjct: 930 VAAFFGA------GLGGGGTFFPGVVLVEVLDALAGLAAGFALAAGLAFGTGGFTF-IRG 772
Query: 114 PTLDMPLIDYDLALLFQPMLMLGISIGVAF 143
LD + L L + + G + +AF
Sbjct: 771 AVLDFA---FALGLAGEAFALAGAATALAF 691
>TC14083 similar to UP|CRTC_RICCO (P93508) Calreticulin precursor, partial
(92%)
Length = 1693
Score = 30.8 bits (68), Expect = 0.47
Identities = 35/118 (29%), Positives = 48/118 (40%), Gaps = 6/118 (5%)
Frame = +1
Query: 107 YNLRLRHPTLDMPLIDYDLALLFQPMLM-LGISIGVAFNVMFADWMV---TILLIILFIG 162
Y + + PT+D P D L P L +GI + W V T+ +L
Sbjct: 973 YQGKWKAPTIDNPDFKDDPDLYVYPKLKYVGIEL----------WQVKSGTLFDNVLITD 1122
Query: 163 TSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPA--DLQDEE 218
A +TW K+ +K AFEEA + E EE+ K PA D +DEE
Sbjct: 1123 DPEYAKQVAEETWGKQKDAEKAAFEEAEKKRE---------EEESKEDPADSDAEDEE 1269
>TC12847 similar to GB|AAM66074.1|21595125|AY088542 endosperm-specific
protein-like protein {Arabidopsis thaliana;} , partial
(11%)
Length = 245
Score = 28.5 bits (62), Expect = 2.3
Identities = 15/31 (48%), Positives = 18/31 (57%)
Frame = -1
Query: 42 EMKFGWRIVVGSIVGFFGAALGSVGGVGGGG 72
E+ G VVG++VG G LG V G GGG
Sbjct: 194 ELGLGEEGVVGAVVGVGGENLGDVVGGDGGG 102
>BP043612
Length = 472
Score = 28.1 bits (61), Expect = 3.1
Identities = 12/31 (38%), Positives = 20/31 (63%)
Frame = +3
Query: 203 SEEDYKSLPADLQDEEVPLLDNIHWKELSVL 233
SEE ++S PA+L EV +W++L+V+
Sbjct: 18 SEEAFESEPAELLSAEVAAFGKSNWEDLTVI 110
>TC8557 similar to UP|Q9LK52 (Q9LK52) Dbj|BAA90629.1, complete
Length = 1064
Score = 27.7 bits (60), Expect = 4.0
Identities = 12/40 (30%), Positives = 22/40 (55%)
Frame = -2
Query: 223 DNIHWKELSVLMYVWVAFLIVQILKTYSKTCSIEYWLLNS 262
D +H+ L ++W +FL+V ++ +S S W LN+
Sbjct: 847 DKVHFMYLCK-NFLWKSFLMVSLMNRFSALASPRAWFLNT 731
>TC9294 homologue to UP|CAE45585 (CAE45585) Coatomer alpha subunit-like
protein, partial (16%)
Length = 1058
Score = 27.3 bits (59), Expect = 5.2
Identities = 20/72 (27%), Positives = 30/72 (40%), Gaps = 1/72 (1%)
Frame = -1
Query: 221 LLDNIHWKELSVLMYVWVAFLIVQILKTYSKTCSIEYWLLNSLQ-VPIAISVTLFEAICL 279
LL ++ W Y WV +IVQ+ C I + S Q +P +S+ CL
Sbjct: 503 LLSSVDWHICRTTDYKWVPEIIVQL-------CGISHISFGSCQHLPCLLSLVFNGRTCL 345
Query: 280 CKGTRVIASRGK 291
+ + SR K
Sbjct: 344 KQPPGKVRSRSK 309
>TC17585 homologue to UP|Q7VBB6 (Q7VBB6) Uncharacterized protein, partial
(5%)
Length = 603
Score = 26.9 bits (58), Expect = 6.8
Identities = 14/39 (35%), Positives = 21/39 (52%)
Frame = -2
Query: 295 WKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFLELG 333
++F L+C G++ ++S LGLG G G L LG
Sbjct: 155 FEFELSSLWCCSGVVVAVLSPKLGLGFGGRKGSWRLNLG 39
>TC9768 similar to GB|AAP68290.1|31711868|BT008851 At3g12156 {Arabidopsis
thaliana;}, partial (51%)
Length = 865
Score = 26.9 bits (58), Expect = 6.8
Identities = 12/46 (26%), Positives = 24/46 (52%)
Frame = +3
Query: 328 LFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLV 373
LFL L + Q+ S + + +++F ++ + D +P+P LV
Sbjct: 21 LFLSLSLLNQIQSLSQSLSLLFLPPFPILPLHVTDMYPLPMILNLV 158
>BP058641
Length = 344
Score = 26.9 bits (58), Expect = 6.8
Identities = 10/25 (40%), Positives = 19/25 (76%)
Frame = -1
Query: 230 LSVLMYVWVAFLIVQILKTYSKTCS 254
L +++ +W+AFL+V +L+T + CS
Sbjct: 236 LRMVLQLWLAFLVVAMLETSACCCS 162
>AU089149
Length = 660
Score = 26.6 bits (57), Expect = 8.9
Identities = 14/27 (51%), Positives = 17/27 (62%)
Frame = -1
Query: 47 WRIVVGSIVGFFGAALGSVGGVGGGGI 73
+ IVV + G G ALG GG GGGG+
Sbjct: 126 YAIVVVDLGGGEGVALG*DGGGGGGGL 46
>TC15734
Length = 656
Score = 26.6 bits (57), Expect = 8.9
Identities = 14/27 (51%), Positives = 17/27 (62%)
Frame = -2
Query: 47 WRIVVGSIVGFFGAALGSVGGVGGGGI 73
+ IVV + G G ALG GG GGGG+
Sbjct: 139 YAIVVVDLGGGEGVALG*DGGGGGGGL 59
>TC16004
Length = 594
Score = 26.6 bits (57), Expect = 8.9
Identities = 21/77 (27%), Positives = 35/77 (45%), Gaps = 4/77 (5%)
Frame = +3
Query: 89 STALSKCMITGAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNV--- 145
ST +S CM + GSTV ++ + T+ + I Y L + F+ L FN+
Sbjct: 111 STVISFCMALTSLGSTVAFDAMVSIATIGL-YIAYALPIFFRVTLARKSFAPGPFNLGRY 287
Query: 146 -MFADWMVTILLIILFI 161
F W+ I ++ + I
Sbjct: 288 GTFVGWISVIWVVTISI 338
>BI418928
Length = 496
Score = 26.6 bits (57), Expect = 8.9
Identities = 16/50 (32%), Positives = 25/50 (50%), Gaps = 5/50 (10%)
Frame = -1
Query: 310 AGMVSGLLGLGGGFILGPLFLELGIPPQVA-----SATSTFAMVFSSSMS 354
+G +S + G GF G G+ P + +STF +VFS+S+S
Sbjct: 337 SGFLSSVFGFSSGFFSGG-----GLSPAASFLGGEETSSTFLLVFSASLS 203
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.142 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,130,006
Number of Sequences: 28460
Number of extensions: 115092
Number of successful extensions: 1030
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 1015
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1026
length of query: 447
length of database: 4,897,600
effective HSP length: 93
effective length of query: 354
effective length of database: 2,250,820
effective search space: 796790280
effective search space used: 796790280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC147964.5


