

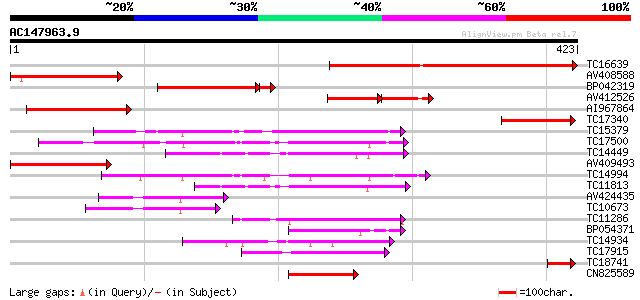
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147963.9 - phase: 0
(423 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC16639 309 6e-85
AV408588 144 2e-35
BP042319 135 1e-33
AV412526 81 3e-29
AI967864 108 2e-24
TC17340 similar to UP|BAC83509 (BAC83509) Protein phosphatase 2C... 98 2e-21
TC15379 UP|Q9ZPL8 (Q9ZPL8) Protein phosphatase type 2C, complete 95 2e-20
TC17500 UP|Q9ZPL9 (Q9ZPL9) Nodule-enhanced protein phosphatase t... 94 6e-20
TC14449 similar to UP|O24078 (O24078) Protein phosphatase 2C, pa... 87 7e-18
AV409493 75 2e-14
TC14994 similar to UP|O82469 (O82469) Protein phosphatase-2C , p... 75 2e-14
TC11813 similar to UP|Q8LJ68 (Q8LJ68) Protein phosphatase 2C-lik... 68 2e-12
AV424435 55 2e-08
TC10673 homologue to UP|Q8S8Z0 (Q8S8Z0) Protein phosphatase 2C, ... 54 4e-08
TC11286 homologue to GB|AAO63448.1|28951049|BT005384 At4g03415 {... 50 7e-07
BP054371 48 3e-06
TC14934 similar to UP|Q9LSN8 (Q9LSN8) Protein phosphatase 2C-lik... 47 8e-06
TC17915 similar to GB|AAM10415.1|20147411|AY093799 AT3g05640/F18... 47 8e-06
TC18741 45 3e-05
CN825589 44 4e-05
>TC16639
Length = 848
Score = 309 bits (791), Expect = 6e-85
Identities = 157/185 (84%), Positives = 166/185 (88%)
Frame = +3
Query: 239 NAGGRLIIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDTTCLVVDII 298
NAGGRLIIASDGIWD LSSDMAAKSCRGVPAELAAKLVVKEALR+RGLKDDTTCLVVDII
Sbjct: 3 NAGGRLIIASDGIWDALSSDMAAKSCRGVPAELAAKLVVKEALRTRGLKDDTTCLVVDII 182
Query: 299 PSDYPVLPMPATPRKKHNVLSSLLFGKKSQNLANKGTNKLSAVGVVEELFEEGSAMLTER 358
PSD+PVLP TPRKK N+L+SLLFGKKSQN NKGTNKLSAVG+VEELFEEGSAMLTER
Sbjct: 183 PSDHPVLP--PTPRKKPNMLTSLLFGKKSQNSTNKGTNKLSAVGIVEELFEEGSAMLTER 356
Query: 359 LGNNVPSDTNSGIHRCAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAM 418
LG + PS+ NSGI RCAVC ADQ DGLSVN++ TPVSKP EGPFLCTNCQKKKDAM
Sbjct: 357 LGKDAPSNVNSGISRCAVCQADQIPSDGLSVNSEPIFTPVSKPLEGPFLCTNCQKKKDAM 536
Query: 419 EGKRS 423
EGKRS
Sbjct: 537 EGKRS 551
>AV408588
Length = 431
Score = 144 bits (364), Expect = 2e-35
Identities = 73/86 (84%), Positives = 78/86 (89%), Gaps = 2/86 (2%)
Frame = +1
Query: 1 MSKAELS--RMKQPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRV 58
M+K+ELS RMK P VPLATLIGRELR+ K EKPFVKYGQAGLAKKGEDYFLI+TDCHRV
Sbjct: 172 MNKSELSSTRMKPPPVPLATLIGRELRHEKVEKPFVKYGQAGLAKKGEDYFLIRTDCHRV 351
Query: 59 PGDSSTSFSVFAILDGHNGISAAIFA 84
PGD ST+FSVFAI DGHNGISAAIFA
Sbjct: 352 PGDPSTAFSVFAIFDGHNGISAAIFA 429
>BP042319
Length = 268
Score = 135 bits (340), Expect(2) = 1e-33
Identities = 66/77 (85%), Positives = 70/77 (90%)
Frame = +3
Query: 111 RALVVAFVKTDMEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVD 170
RALV FVKTD EFQ +GETSGTTATFVI+D WTVTVASVGDSRCILDTQGG V+ LTVD
Sbjct: 3 RALVAGFVKTDKEFQSRGETSGTTATFVIVDRWTVTVASVGDSRCILDTQGGAVTTLTVD 182
Query: 171 HRLEENVEERERVTASG 187
HRLEEN+EERERVTASG
Sbjct: 183 HRLEENIEERERVTASG 233
Score = 24.3 bits (51), Expect(2) = 1e-33
Identities = 9/12 (75%), Positives = 11/12 (91%)
Frame = +1
Query: 187 GGEVGRLNVFGG 198
GGEVGRL++ GG
Sbjct: 232 GGEVGRLSIVGG 267
>AV412526
Length = 325
Score = 80.9 bits (198), Expect(2) = 3e-29
Identities = 40/41 (97%), Positives = 40/41 (97%)
Frame = +1
Query: 238 SNAGGRLIIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVK 278
SNAGGRLIIASDGIWD LSSDMAAKSCRGVPAELAAKLVVK
Sbjct: 1 SNAGGRLIIASDGIWDALSSDMAAKSCRGVPAELAAKLVVK 123
Score = 64.3 bits (155), Expect(2) = 3e-29
Identities = 32/39 (82%), Positives = 35/39 (89%)
Frame = +2
Query: 278 KEALRSRGLKDDTTCLVVDIIPSDYPVLPMPATPRKKHN 316
+EALR+RGLKDDTTCLVVDIIPSD+PVL P TPRKK N
Sbjct: 215 QEALRTRGLKDDTTCLVVDIIPSDHPVL--PPTPRKKPN 325
>AI967864
Length = 405
Score = 108 bits (269), Expect = 2e-24
Identities = 50/79 (63%), Positives = 66/79 (83%)
Frame = +3
Query: 13 LVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAIL 72
LVPLA+L+ RE+++ K EKP +++G A +KKGEDYFLIKTDC RVPG+SS+SFSVFAI
Sbjct: 165 LVPLASLLKREMKSEKMEKPTLRFGHAAQSKKGEDYFLIKTDCQRVPGNSSSSFSVFAIF 344
Query: 73 DGHNGISAAIFAKENIINN 91
DGHNG +AAI+ +E I+ +
Sbjct: 345 DGHNGYAAAIYYQEPIVQS 401
>TC17340 similar to UP|BAC83509 (BAC83509) Protein phosphatase 2C-like,
partial (8%)
Length = 749
Score = 98.2 bits (243), Expect = 2e-21
Identities = 43/55 (78%), Positives = 47/55 (85%)
Frame = +1
Query: 368 NSGIHRCAVCLADQPSGDGLSVNNDHFITPVSKPWEGPFLCTNCQKKKDAMEGKR 422
NSGI RCAVC DQP GDGLSVN+ F +P S+PWEGPFLCTNC+KKKDAMEGKR
Sbjct: 4 NSGIVRCAVCQVDQPPGDGLSVNSGPFFSPASQPWEGPFLCTNCRKKKDAMEGKR 168
>TC15379 UP|Q9ZPL8 (Q9ZPL8) Protein phosphatase type 2C, complete
Length = 1338
Score = 95.1 bits (235), Expect = 2e-20
Identities = 72/238 (30%), Positives = 118/238 (49%), Gaps = 5/238 (2%)
Frame = +1
Query: 63 STSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDM 122
+ +FAI DGH+G + + + N+ +N++ + W + + A+ A+V TD
Sbjct: 403 NNELGLFAIFDGHSGHNVPDYLQSNLFDNILK------EPDFWTKPV-EAVKKAYVDTDS 561
Query: 123 EFQKK----GETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVE 178
+K G T T ++I+ + VA++GDSR +L G + L +VDH E
Sbjct: 562 TILEKSGELGRGGSTAVTAILINCQKLVVANLGDSRAVLCKNGEAIPL-SVDH---EPAT 729
Query: 179 ERERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLS 238
E E + GG F N G + G L +SR+ GD + +++ PHV +
Sbjct: 730 ESEDIRNRGG-------FVSNFPGDVPRVDGQLAVSRAFGDKSLKKHLSSEPHVTVELID 888
Query: 239 NAGGRLIIASDGIWDTLSSDMAAKSCRGV-PAELAAKLVVKEALRSRGLKDDTTCLVV 295
+ +I+ASDG+W +S+ A + R V A AAK + +EAL+ R DD +C+VV
Sbjct: 889 DDAEFIILASDGLWKVMSNQEAVDAIRNVKDARSAAKNLTEEALK-RNSSDDISCVVV 1059
>TC17500 UP|Q9ZPL9 (Q9ZPL9) Nodule-enhanced protein phosphatase type 2C,
complete
Length = 1325
Score = 93.6 bits (231), Expect = 6e-20
Identities = 84/295 (28%), Positives = 130/295 (43%), Gaps = 19/295 (6%)
Frame = +3
Query: 22 RELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISAA 81
RE + P+ G K+ ED ++T C T FA+ DGH G A
Sbjct: 363 RERKRDDGVLPYGSVSVVGSRKEMEDAVSVETGC-------VTKCDYFAVFDGHGGAQVA 521
Query: 82 IFAKENIINNVMSAIPQ---GVSREEWLQALPRALVVAFVKTDMEFQKKG--ETSGTTAT 136
+E + V + + GV +W + + F D E T G+TA
Sbjct: 522 EACRERLYRLVAEEVERCGNGVEEVDWEEVMEGC----FRNMDGEVAGNAALRTVGSTAV 689
Query: 137 FVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVF 196
++ V +A+ GD R +L G V L + DH+ + +E R+ +GG+V N
Sbjct: 690 VAVVAAAEVVIANCGDCRAVLGRGGEAVDLSS-DHK-PDRPDELMRIEEAGGKVINWN-- 857
Query: 197 GGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLS 256
G V G L SRSIGD + Y++ P V K S+ LI+ASDG+WD +S
Sbjct: 858 -GQRV------LGVLATSRSIGDQYLRPYVISKPEVTVTKRSSKDEFLILASDGLWDVIS 1016
Query: 257 SDMAAKSCRGV--------------PAELAAKLVVKEALRSRGLKDDTTCLVVDI 297
S+MA + R A AA L+ + AL ++G +D+T+ +V+++
Sbjct: 1017SEMACQVVRKCLNGQIRRICNENQSRASEAATLLAEIAL-AKGSRDNTSVIVIEL 1178
>TC14449 similar to UP|O24078 (O24078) Protein phosphatase 2C, partial (63%)
Length = 1000
Score = 86.7 bits (213), Expect = 7e-18
Identities = 60/187 (32%), Positives = 94/187 (50%), Gaps = 6/187 (3%)
Frame = +3
Query: 117 FVKTDMEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEEN 176
++ TD +F K+ G+ +I + V++ GD R ++ ++GGV LT DHR
Sbjct: 180 YLNTDSDFLKEDLHGGSCCVTALIRNGNLVVSNAGDCRAVI-SRGGVAEALTTDHRPSRE 356
Query: 177 VEERERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVK 236
+ERER+ GG V G R G L +SRSIGD + +++ P K +K
Sbjct: 357 -DERERIETLGGYVDLCR-------GVWRI-QGSLAVSRSIGDRHLKQWVTAEPETKVIK 509
Query: 237 LSNAGGRLIIASDGIWDTLSS----DMAAKSCRG--VPAELAAKLVVKEALRSRGLKDDT 290
+ LI+ASDG+WD +S+ D+A C G P L A + + SRG DDT
Sbjct: 510 IEPEHDLLILASDGLWDKVSNQEAVDLARPLCVGNNKPQSLLACKKLVDLACSRGSMDDT 689
Query: 291 TCLVVDI 297
+ +++ +
Sbjct: 690 SVMLIKL 710
>AV409493
Length = 426
Score = 75.5 bits (184), Expect = 2e-14
Identities = 34/76 (44%), Positives = 50/76 (65%)
Frame = +3
Query: 1 MSKAELSRMKQPLVPLATLIGRELRNGKTEKPFVKYGQAGLAKKGEDYFLIKTDCHRVPG 60
MS + VPL+ L+ REL + + EKP + +GQAG +KKGED+ L+K +C R+ G
Sbjct: 198 MSTTTRNEQHHQTVPLSVLLKRELASERIEKPEIVHGQAGQSKKGEDFTLLKAECQRMVG 377
Query: 61 DSSTSFSVFAILDGHN 76
D +++SVF + DGHN
Sbjct: 378 DGVSTYSVFGLFDGHN 425
>TC14994 similar to UP|O82469 (O82469) Protein phosphatase-2C , partial
(89%)
Length = 1672
Score = 75.5 bits (184), Expect = 2e-14
Identities = 79/265 (29%), Positives = 121/265 (44%), Gaps = 19/265 (7%)
Frame = +3
Query: 69 FAILDGHNGISAAIFAKENIINNVMSAI--PQGVSREE-WLQALPRALVVAFVKTDMEFQ 125
+ + DGH G AA + ++N+I + PQ ++ +LQ + +L AF+ D
Sbjct: 615 YGVFDGHGGPEAAAYIRKNVIKFFFEDVSFPQTSEVDKVFLQEVENSLRKAFLLADSALA 794
Query: 126 KK---GETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERER 182
+SGTTA I G + VA+ GD R +L +G + + + DHR ER R
Sbjct: 795 DDCSVNTSSGTTALTAFIFGRLLMVANAGDCRAVLSRKGEAIDM-SEDHR-PIYPSERRR 968
Query: 183 VTASGG--EVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVG------EYIVPIPHVKQ 234
V GG E G LN G L ++R++GD D+ ++ P +Q
Sbjct: 969 VEDLGGYIEDGYLN--------------GVLSVTRALGDWDMKLPKGAPSPLIAEPEFRQ 1106
Query: 235 VKLSNAGGRLIIASDGIWDTLSSDMAAKSCR-GV----PAELAAKLVVKEALRSRGLKDD 289
+ L+ LII DGIWD +SS A R G+ E A+ +V EALR D+
Sbjct: 1107MVLTEDDEFLIIGCDGIWDVMSSQHAVSLVRKGLRRHDDPERCARDLVMEALRLNTF-DN 1283
Query: 290 TTCLVVDIIPSDYPVLPMPATPRKK 314
T ++V D+ P+ P++K
Sbjct: 1284LTVIIVCFSSLDHRE-SEPSPPQRK 1355
>TC11813 similar to UP|Q8LJ68 (Q8LJ68) Protein phosphatase 2C-like protein,
partial (36%)
Length = 917
Score = 68.2 bits (165), Expect = 2e-12
Identities = 57/182 (31%), Positives = 89/182 (48%), Gaps = 21/182 (11%)
Frame = +1
Query: 139 IIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGG 198
+I + V++ GDSR +L +G L+VDH+ +E R+ A+GG+V + N G
Sbjct: 16 LISSSHIIVSNCGDSRAVL-CRGKEPLALSVDHKPNRE-DEYARIEAAGGKVIQWN---G 180
Query: 199 NEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSD 258
+ V G L +SRSIGD + +I+P P V+ + + LI+ASDG+WD ++++
Sbjct: 181 HRVF------GVLAMSRSIGDRYLKPWIIPEPEVRFLPRAKEDECLILASDGLWDVMTNE 342
Query: 259 MAAKSCR--------------------GV-PAELAAKLVVKEALRSRGLKDDTTCLVVDI 297
A R GV PA AA + +G KD+ T +VVD+
Sbjct: 343 EACDLARKRILLWYKKNGFELSSERGEGVDPAAQAAAEFLSNRALPKGSKDNITVIVVDL 522
Query: 298 IP 299
P
Sbjct: 523 KP 528
>AV424435
Length = 280
Score = 55.5 bits (132), Expect = 2e-08
Identities = 34/99 (34%), Positives = 53/99 (53%), Gaps = 2/99 (2%)
Frame = +1
Query: 67 SVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQK 126
S + + DGH G SAA F ++++ + V ++ L + + +F++TD EF K
Sbjct: 4 SFYGVFDGHGGKSAAQFVRDHLPRVI-------VEDADFPLELEKVVTKSFLETDAEFAK 162
Query: 127 --KGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGV 163
E+SGTTA II G ++ VA+ GD R +L G V
Sbjct: 163 TCSSESSGTTALTAIILGRSLLVANAGDCRAVLSRSGAV 279
>TC10673 homologue to UP|Q8S8Z0 (Q8S8Z0) Protein phosphatase 2C, partial
(48%)
Length = 669
Score = 54.3 bits (129), Expect = 4e-08
Identities = 31/105 (29%), Positives = 55/105 (51%), Gaps = 4/105 (3%)
Frame = +3
Query: 57 RVPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVA 116
R+ G +F + DGH G AA + K+N+ +N+ +S +++ A+ A
Sbjct: 369 RIDGVEGEVVGLFGVFDGHGGARAAEYVKQNLFSNL-------ISHPKFISDTKSAIADA 527
Query: 117 FVKTDMEFQK----KGETSGTTATFVIIDGWTVTVASVGDSRCIL 157
+ TD EF K + +G+TA+ I+ G + VA+VGDS+ ++
Sbjct: 528 YNHTDSEFLKSENNQNRDAGSTASTAILVGDRLLVANVGDSKAVI 662
>TC11286 homologue to GB|AAO63448.1|28951049|BT005384 At4g03415 {Arabidopsis
thaliana;}, partial (46%)
Length = 996
Score = 50.1 bits (118), Expect = 7e-07
Identities = 45/140 (32%), Positives = 66/140 (47%), Gaps = 11/140 (7%)
Frame = +3
Query: 167 LTVDHRLEENVEERERVTASGGEVGRLNVFGGNEVGPLRCW-----PGGLCLSRSIGDTD 221
LTVD + + E ER+ G V L +E R W GL ++R+ GD
Sbjct: 30 LTVDLK-PDLPREAERIKRCKGRVFALQ----DEPEVPRVWLPFDDAPGLAMARAFGDFC 194
Query: 222 VGEY-IVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMAAKSCRGVPAEL-AAKLVVKE 279
+ EY ++ IP +L++ ++IASDG+WD LS++ + P AA+ VV
Sbjct: 195 LKEYGVISIPEFSHRQLTDRDQFIVIASDGVWDVLSNEEVVEIVSSSPTRASAARTVVNS 374
Query: 280 ALRSRGLKDDTT----CLVV 295
A R LK T+ C VV
Sbjct: 375 AAREWKLKYPTSRMDDCAVV 434
>BP054371
Length = 567
Score = 48.1 bits (113), Expect = 3e-06
Identities = 32/90 (35%), Positives = 52/90 (57%), Gaps = 3/90 (3%)
Frame = -2
Query: 209 GGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMA---AKSCR 265
G L +SR+ GD ++ ++ P ++ ++ + LI+ASDG+W +++ A AK +
Sbjct: 554 GQLAVSRAFGDQNLKSHLRSDPDIRHDEIDSDTEFLILASDGLWKVMANPEAVDIAKKIQ 375
Query: 266 GVPAELAAKLVVKEALRSRGLKDDTTCLVV 295
P AAK + EAL +R KDD +CLVV
Sbjct: 374 DPPK--AAKPLTAEAL-NRESKDDISCLVV 294
>TC14934 similar to UP|Q9LSN8 (Q9LSN8) Protein phosphatase 2C-like protein,
partial (57%)
Length = 932
Score = 46.6 bits (109), Expect = 8e-06
Identities = 52/186 (27%), Positives = 86/186 (45%), Gaps = 28/186 (15%)
Frame = +1
Query: 130 TSGTTATFVIIDGWTVTVASVGDSRCILDTQ----GGVVSL-LTVDHR--LEENVEERER 182
T GT +I T+ VA++GDSRC+L + GGV ++ L+ +H LE+ +E +
Sbjct: 4 TRGTCCLVGVIFQQTLFVANLGDSRCVLGKKVGNTGGVAAIQLSTEHNANLEDIRQELKE 183
Query: 183 VTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVG-----------EYIVPIPH 231
+ ++ L + G R G + +SRSIGD+ + ++ +P P
Sbjct: 184 LHPDDPQIVVL------KHGVWRV-KGIIQVSRSIGDSYMKHAQFNREPINPKFRLPEPM 342
Query: 232 VKQVKLSN----------AGGRLIIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEAL 281
+ +N + LI ASDG+W+ LS++ A P +AK +VK AL
Sbjct: 343 NMPIMSANPTILSHPLQPSDSFLIFASDGLWEHLSNEQAVDIVHSNPRAGSAKRLVKAAL 522
Query: 282 RSRGLK 287
+ K
Sbjct: 523 QEAARK 540
>TC17915 similar to GB|AAM10415.1|20147411|AY093799 AT3g05640/F18C1_9
{Arabidopsis thaliana;}, partial (37%)
Length = 506
Score = 46.6 bits (109), Expect = 8e-06
Identities = 34/112 (30%), Positives = 57/112 (50%), Gaps = 2/112 (1%)
Frame = +2
Query: 174 EENVEERERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEY-IVPIPHV 232
E +E + RV E G V +E P GL +SR+ GD V EY ++ +P V
Sbjct: 2 ERILEHQGRVFCLDDEPGVHRVCLPDEESP------GLAMSRAFGDYCVKEYGLISVPEV 163
Query: 233 KQVKLSNAGGRLIIASDGIWDTLSSDMAAKSCRGVPAEL-AAKLVVKEALRS 283
Q +++ +++A+DG+WD +S++ A +AK +VK A+R+
Sbjct: 164 TQRNITSKDQFVVLATDGVWDVISNEEAVDIVSSTADRTDSAKRLVKCAMRA 319
>TC18741
Length = 669
Score = 44.7 bits (104), Expect = 3e-05
Identities = 16/21 (76%), Positives = 21/21 (99%)
Frame = +3
Query: 402 WEGPFLCTNCQKKKDAMEGKR 422
W+GPFLC++CQ+KK+AMEGKR
Sbjct: 3 WDGPFLCSSCQEKKEAMEGKR 65
>CN825589
Length = 544
Score = 44.3 bits (103), Expect = 4e-05
Identities = 21/52 (40%), Positives = 32/52 (61%)
Frame = +3
Query: 209 GGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMA 260
G L +SR+IGD + Y++ P V ++ LI+ASDG+WD +S+D A
Sbjct: 51 GVLAMSRAIGDNYLKPYVISEPEVTVTDRTDEDECLILASDGLWDVVSNDTA 206
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,820,726
Number of Sequences: 28460
Number of extensions: 89508
Number of successful extensions: 375
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 367
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 369
length of query: 423
length of database: 4,897,600
effective HSP length: 93
effective length of query: 330
effective length of database: 2,250,820
effective search space: 742770600
effective search space used: 742770600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC147963.9


