

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147960.1 - phase: 1 /pseudo
(451 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
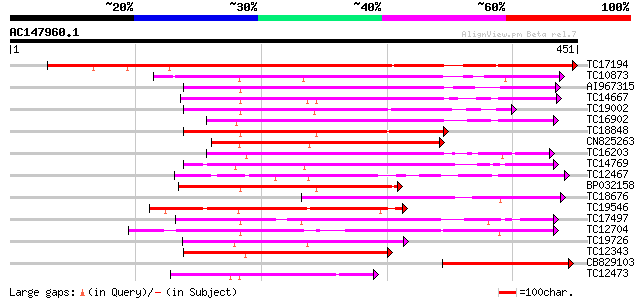
Score E
Sequences producing significant alignments: (bits) Value
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 411 e-115
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 199 1e-51
AI967315 174 2e-44
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 169 1e-42
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 169 1e-42
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 167 4e-42
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 163 5e-41
CN825263 160 4e-40
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 158 2e-39
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 157 4e-39
TC12467 UP|CAE02597 (CAE02597) Nod-factor receptor 5, complete 155 2e-38
BP032158 149 7e-37
TC18676 similar to UP|Q8RYS1 (Q8RYS1) Protein kinase-like, parti... 148 2e-36
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 146 8e-36
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 142 9e-35
TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3,... 135 1e-32
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 132 9e-32
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 132 2e-31
CB829103 130 4e-31
TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protei... 122 9e-29
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 411 bits (1056), Expect = e-115
Identities = 238/466 (51%), Positives = 304/466 (65%), Gaps = 45/466 (9%)
Frame = +1
Query: 31 REFLYFLYLPFCP*ATLESIAKDTMIEIELLKRYN--VNFSQGSGLVYIRGKDGNGLYVP 88
+++ F+ P P TL+ IA + ++ L++ +N VNFS+ SG+ +I G+ NG+YVP
Sbjct: 595 KDYGLFITYPIRPGDTLQDIANQSSLDAGLIQSFNPSVNFSKDSGIAFIPGRYKNGVYVP 774
Query: 89 LPPK---------------------------YFR-KKNGEEESKFPPEDSMTPSTKDVDK 120
L + Y R +K EE++K P + SM ST+D +
Sbjct: 775 LYHRTAGLASGAAVGISIAGTFVLLLLAFCMYVRYQKKEEEKAKLPTDISMALSTQDGNA 954
Query: 121 DTNDD--------------NGSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVY 166
++ + G I V KS EFSY+ELA AT+NFSL KIGQGGFG VY
Sbjct: 955 SSSAEYETSGSSGPGTASATGLTSIMVAKSMEFSYQELAKATNNFSLDNKIGQGGFGAVY 1134
Query: 167 YGELRGQKIAIKKMKMQATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGN 226
Y ELRG+K AIKKM +QA+ EFL ELKVLT VHH NLV LIGYCVEG LFLVYE+++NGN
Sbjct: 1135 YAELRGKKTAIKKMDVQASTEFLCELKVLTHVHHLNLVRLIGYCVEGSLFLVYEHIDNGN 1314
Query: 227 LSQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKV 286
L Q+LH S KEP+ S+R++IALD ARGLEYIH+H+VPVYIHRD+KS NIL+++N GKV
Sbjct: 1315 LGQYLHGSGKEPLPWSSRVQIALDAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKV 1494
Query: 287 ADFGLTKLTDAANSADNTVHVAGTFGYMPPENA-YGRISRKIDVYAFGVVLYELISAKAA 345
ADFGLTKL + NS T + GTFGYMPPE A YG IS KIDVYAFGVVL+ELISAK A
Sbjct: 1495 ADFGLTKLIEVGNSTLQT-RLVGTFGYMPPEYAQYGDISPKIDVYAFGVVLFELISAKNA 1671
Query: 346 VIKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSI 405
V +KT E + E K LVALF+E +N++ C D LRKLVDPRLG NY I
Sbjct: 1672 V--------------LKTGELVAESKGLVALFEEALNKSDPC-DALRKLVDPRLGENYPI 1806
Query: 406 DSISKMAKLAKACINRDPKQRPTMRDVVVSLMELNSSIDNKNSTGS 451
DS+ K+A+L +AC +P RP+MR +VV+LM L+S ++ + S
Sbjct: 1807 DSVLKIAQLGRACTRDNPLLRPSMRSLVVALMTLSSLTEDCDDESS 1944
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 199 bits (505), Expect = 1e-51
Identities = 128/338 (37%), Positives = 185/338 (53%), Gaps = 11/338 (3%)
Frame = +1
Query: 115 TKDVDKDTNDDNGSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELR-GQ 173
TK K + NGS + F + ELA+AT NF A IG+GGFG+VY G L G+
Sbjct: 346 TKGKGKGVSSSNGSSN-GKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGE 522
Query: 174 KIAIKKMK---MQATREFLSELKVLTSVHHWNLVHLIGYCVEGFL-FLVYEYMENGNLSQ 229
+A+K++ Q +EF+ E+ +L+ +HH NLV LIGYC +G LVYEYM G+L
Sbjct: 523 AVAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLED 702
Query: 230 HLH--NSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVA 287
HL + +KEP+ STRMK+A+ ARGLEY+H + P I+RD+KS NILL+ F K++
Sbjct: 703 HLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLS 882
Query: 288 DFGLTKLTDAANSADNTVHVAGTFGYMPPENAY-GRISRKIDVYAFGVVLYELISAKAAV 346
DFGL KL ++ + V GT+GY PE A G+++ K D+Y+FGVVL EL++ + A
Sbjct: 883 DFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRA- 1059
Query: 347 IKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRK---LVDPRLGYNY 403
I T+ E +++ D R+ +VDP L +
Sbjct: 1060--------------IDTSRRPGE--------QNLVSWARPYFSDRRRFGHMVDPLLQGRF 1173
Query: 404 SIDSISKMAKLAKACINRDPKQRPTMRDVVVSLMELNS 441
+ + + C+ PK RP + D+VV+L L S
Sbjct: 1174PSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLAS 1287
>AI967315
Length = 1308
Score = 174 bits (442), Expect = 2e-44
Identities = 106/307 (34%), Positives = 165/307 (53%), Gaps = 7/307 (2%)
Frame = +1
Query: 139 FSYEELANATDNFSLAKKIGQGGFGEVYYGELR-GQKIAIKKMKM-----QATREFLSEL 192
FSYEEL +AT+ FS +G+GG+ EVY G L G +IA+K++ + +EFL+E+
Sbjct: 112 FSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEI 291
Query: 193 KVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVA 252
+ V H N++ L+G C++ L+LV+E G+++ +H+ + P+ TR KI L A
Sbjct: 292 GTIGHVCHSNVMPLLGCCIDNGLYLVFELSTVGSVASLIHDEKMAPLDWKTRYKIVLGTA 471
Query: 253 RGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFG 312
RGL Y+H IHRDIK+ NILL E+F +++DFGL K + + + + GTFG
Sbjct: 472 RGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIEGTFG 651
Query: 313 YMPPE-NAYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYK 371
++ PE +G + K DV+AFGV L E+IS + V + +SL + ++
Sbjct: 652 HLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPV------DGSHQSLHTWAKPILSKW- 810
Query: 372 SLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRD 431
++ KLVDPRL Y + +++A A CI RPTM +
Sbjct: 811 ------------------EIEKLVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSE 936
Query: 432 VVVSLME 438
V+ + E
Sbjct: 937 VLEVMEE 957
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 169 bits (427), Expect = 1e-42
Identities = 112/317 (35%), Positives = 172/317 (53%), Gaps = 14/317 (4%)
Frame = +2
Query: 137 PEFSYEELANATDNFSLAKKIGQGGFGEVYYGELR-GQKIAIKKMKM----QATREFLSE 191
P S +EL TDNF IG+G +G VYY L G +A+KK+ + + EFL++
Sbjct: 317 PALSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNEFLTQ 496
Query: 192 LKVLTSVHHWNLVHLIGYCVEGFL-FLVYEYMENGNLSQHLHNSE-----KEPMTLS--T 243
+ +++ + + N V L GYCVEG L L YE+ G+L LH + + TL
Sbjct: 497 VSMVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQ 676
Query: 244 RMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADN 303
R++IA+D ARGLEY+H+ P IHRDI+S N+L+ E++ K+ADF L+ + +
Sbjct: 677 RVRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLH 856
Query: 304 TVHVAGTFGYMPPENAY-GRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIK 362
+ V GTFGY PE A G++++K DVY+FGVVL EL++ + V D T +
Sbjct: 857 STRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPV---DHTMPRGQ----- 1012
Query: 363 TNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRD 422
+SLV +++ D +++ VDP+L Y ++K+A +A C+ +
Sbjct: 1013--------QSLVTWATPRLSE-----DKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYE 1153
Query: 423 PKQRPTMRDVVVSLMEL 439
+ RP M VV +L L
Sbjct: 1154AEFRPNMSIVVKALQPL 1204
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 169 bits (427), Expect = 1e-42
Identities = 106/274 (38%), Positives = 151/274 (54%), Gaps = 9/274 (3%)
Frame = +1
Query: 139 FSYEELANATDNFSLAKKIGQGGFGEVYYGEL-RGQKIAIKKMKM---QATREFLSELKV 194
FS +EL +AT+NF+ K+G+GGFG VY+G+L G +IA+K++K+ +A EF E+++
Sbjct: 28 FSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEI 207
Query: 195 LTSVHHWNLVHLIGYCVEGF-LFLVYEYMENGNLSQHLHNSEKEPMTL--STRMKIALDV 251
L V H NL+ L GYC EG +VY+YM N +L HLH L + RM IA+
Sbjct: 208 LARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGS 387
Query: 252 ARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKL-TDAANSADNTVHVAGT 310
A G+ Y+H + P IHRDIK+ N+LL+ +F +VADFG KL D A T V GT
Sbjct: 388 AEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHV--TTRVKGT 561
Query: 311 FGYMPPENA-YGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDE 369
GY+ PE A G+ + DV++FG++L EL S K + K+ T SI++
Sbjct: 562 LGYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSST----------VKRSIND 711
Query: 370 YKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNY 403
+ +A C + DPRL Y
Sbjct: 712 WALPLA-----------CAKKFTEFADPRLNGEY 780
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 167 bits (422), Expect = 4e-42
Identities = 103/287 (35%), Positives = 163/287 (55%), Gaps = 7/287 (2%)
Frame = +3
Query: 157 IGQGGFGEVYYGELRG-QKIAIKK---MKMQATREFLSELKVLTSVHHWNLVHLIGYCVE 212
+G GGFG+VYYGE+ G K+AIK+ + Q EF +E+++L+ + H +LV LIGYC E
Sbjct: 15 LGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYCEE 194
Query: 213 GF-LFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDI 271
+ LVY++M G L +HL+ ++K P+ R++I + ARGL Y+H + IHRD+
Sbjct: 195 NTEMILVYDHMAYGTLREHLYKTQKPPLPWKQRLEICIGAARGLHYLHTGAKYTIIHRDV 374
Query: 272 KSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGYMPPEN-AYGRISRKIDVY 330
K+ NILL+E + KV+DFGL+K ++ + V G+FGY+ PE +++ K DVY
Sbjct: 375 KTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLTDKSDVY 554
Query: 331 AFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESI-DEYKSLVALFDEVMNQTGDCID 389
+FGVVL+E++ A+ A+ N S+ E SL N+
Sbjct: 555 SFGVVLFEILCARPAL-----------------NPSLAKEQVSLAEWASHCYNK-----G 668
Query: 390 DLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVVVSL 436
L +++DP L + + K A+ A C++ +RP+M DV+ +L
Sbjct: 669 ILDQILDPYLKGKIAPECFKKFAETAMKCVSDQGIERPSMGDVLWNL 809
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 163 bits (413), Expect = 5e-41
Identities = 97/219 (44%), Positives = 133/219 (60%), Gaps = 8/219 (3%)
Frame = +2
Query: 139 FSYEELANATDNFSLAKKIGQGGFGEVYYGELR-GQKIAIKKMKM---QATREFLSELKV 194
F+Y+EL AT FS K+G+GGFG VY+G G +IA+KK+K +A EF E++V
Sbjct: 266 FTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 445
Query: 195 LTSVHHWNLVHLIGYCV-EGFLFLVYEYMENGNLSQHLHNSEKEPMTLS--TRMKIALDV 251
L V H NL+ L GYCV + +VY+YM N +L HLH + L+ RMKIA+
Sbjct: 446 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIGS 625
Query: 252 ARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTF 311
A G+ Y+H P IHRDIK+ N+LLN +F VADFG KL S T V GT
Sbjct: 626 AEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVS-HMTTRVKGTL 802
Query: 312 GYMPPENA-YGRISRKIDVYAFGVVLYELISAKAAVIKI 349
GY+ PE A +G++S DVY+FG++L EL++ + + K+
Sbjct: 803 GYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKL 919
>CN825263
Length = 663
Score = 160 bits (405), Expect = 4e-40
Identities = 92/194 (47%), Positives = 125/194 (64%), Gaps = 8/194 (4%)
Frame = +1
Query: 161 GFGEVYYGELR-GQKIAIKKMK---MQATREFLSELKVLTSVHHWNLVHLIGYCVEGFL- 215
GFG VY G L G+ +A+K +K + REFL+E+++L+ +HH NLV LIG C+E
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTR 180
Query: 216 FLVYEYMENGNLSQHLHNSEKE--PMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKS 273
L+YE + NG++ HLH ++KE P+ + RMKIAL ARGL Y+H+ S P IHRD KS
Sbjct: 181 CLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFKS 360
Query: 274 DNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGYMPPENAY-GRISRKIDVYAF 332
NILL +FT KV+DFGL + + + HV GTFGY+ PE A G + K DVY++
Sbjct: 361 SNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYSY 540
Query: 333 GVVLYELISAKAAV 346
GVVL EL++ V
Sbjct: 541 GVVLLELLTGTKPV 582
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 158 bits (400), Expect = 2e-39
Identities = 104/286 (36%), Positives = 157/286 (54%), Gaps = 9/286 (3%)
Frame = +2
Query: 157 IGQGGFGEVYYGEL-RGQKIAIKKMKMQATRE----FLSELKVLTSVHHWNLVHLIGYCV 211
IG+GG G VY G + G +AIK++ Q + F +E++ L + H N++ L+GY
Sbjct: 2210 IGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVS 2389
Query: 212 -EGFLFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRD 270
+ L+YEYM NG+L + LH ++ + R KIA++ ARGL Y+H P+ IHRD
Sbjct: 2390 NKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRD 2569
Query: 271 IKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGYMPPENAYG-RISRKIDV 329
+KS+NILL+ +F VADFGL K ++ + +AG++GY+ PE AY ++ K DV
Sbjct: 2570 VKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDV 2749
Query: 330 YAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCID 389
Y+FGVVL ELI + V EF + +D +V ++ M++ D
Sbjct: 2750 YSFGVVLLELIIGRKPV-----GEF---------GDGVD----IVGWVNKTMSELSQPSD 2875
Query: 390 D--LRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVV 433
+ +VDPRL Y + S+ M +A C+ RPTMR+VV
Sbjct: 2876 TALVLAVVDPRLS-GYPLTSVIHMFNIAMMCVKEMGPARPTMREVV 3010
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 157 bits (396), Expect = 4e-39
Identities = 110/307 (35%), Positives = 166/307 (53%), Gaps = 9/307 (2%)
Frame = +3
Query: 139 FSYEELANATDNFSLAKKIGQGGFGEVYYGELR-GQKIAIK---KMKMQATREFLSELKV 194
F+ E + AT+ + IG+GGFG VY G L GQ++A+K Q TREF +EL +
Sbjct: 2133 FTLEYIEVATERYKTL--IGEGGFGSVYRGTLNDGQEVAVKVRSSTSTQGTREFDNELNL 2306
Query: 195 LTSVHHWNLVHLIGYCVEGFL-FLVYEYMENGNLSQHLHN--SEKEPMTLSTRMKIALDV 251
L+++ H NLV L+GYC E LVY +M NG+L L+ ++++ + TR+ IAL
Sbjct: 2307 LSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALGA 2486
Query: 252 ARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTF 311
ARGL Y+H IHRDIKS NILL+ + KVADFG +K + ++ V GT
Sbjct: 2487 ARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTA 2666
Query: 312 GYMPPE-NAYGRISRKIDVYAFGVVLYELISAKAAV-IKIDKTEFEFKSLEIKTNESIDE 369
GY+ PE ++S K DV++FGVVL E++S + + IK +TE+
Sbjct: 2667 GYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEW--------------- 2801
Query: 370 YKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTM 429
SLV + G +D+ +VDP + Y +++ ++ ++A C+ RP+M
Sbjct: 2802 --SLVEWATPYIR--GSKVDE---IVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSM 2960
Query: 430 RDVVVSL 436
+V L
Sbjct: 2961 VAIVREL 2981
>TC12467 UP|CAE02597 (CAE02597) Nod-factor receptor 5, complete
Length = 2292
Score = 155 bits (391), Expect = 2e-38
Identities = 110/320 (34%), Positives = 164/320 (50%), Gaps = 6/320 (1%)
Frame = +3
Query: 132 WVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAIKKMKMQATREFLSE 191
+V K + +E+ AT +FS K+G+ VY + G+ +A+KK+K E E
Sbjct: 1029 YVSKPNVYEIDEIMEATKDFSDECKVGES----VYKANIEGRVVAVKKIKEGGANE---E 1187
Query: 192 LKVLTSVHHWNLVHLIGYC--VEGFLFLVYEYMENGNLSQHLHNSEK---EPMTLSTRMK 246
LK+L V+H NLV L+G +G FLVYEY ENG+L++ L + +T S R+
Sbjct: 1188 LKILQKVNHGNLVKLMGVSSGYDGNCFLVYEYAENGSLAEWLFSKSSGTPNSLTWSQRIS 1367
Query: 247 IALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVH 306
IA+DVA GL+Y+H+H+ P IHRDI + NILL+ NF K+A+F + +
Sbjct: 1368 IAVDVAVGLQYMHEHTYPRIIHRDITTSNILLDSNFKAKIANFAMAR------------- 1508
Query: 307 VAGTFGYMPPENAYGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTN-E 365
T MP KIDV+AFGV+L EL++ + K++ K N E
Sbjct: 1509 -TSTNPMMP----------KIDVFAFGVLLIELLTGR-------------KAMTTKENGE 1616
Query: 366 SIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQ 425
+ +K + +FD N+ + +RK +DP L Y ID+ +A LA C
Sbjct: 1617 VVMLWKDMWEIFDIEENRE----ERIRKWMDPNLESFYHIDNALSLASLAVNCTADKSLS 1784
Query: 426 RPTMRDVVVSLMELNSSIDN 445
RP+M ++V+SL L N
Sbjct: 1785 RPSMAEIVLSLSFLTQQSSN 1844
>BP032158
Length = 555
Score = 149 bits (377), Expect = 7e-37
Identities = 82/185 (44%), Positives = 124/185 (66%), Gaps = 7/185 (3%)
Frame = +2
Query: 135 KSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGEL-RGQKIAIKKMKM---QATREFLS 190
K+ FS ++ AT+NF A KIG+GGFG VY G L G IA+K++ Q REF++
Sbjct: 2 KTGYFSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSSKSKQGNREFIN 181
Query: 191 ELKVLTSVHHWNLVHLIGYCVEGF-LFLVYEYMENGNLSQHLHNSEKEPMTLS--TRMKI 247
E+ +++++ H NLV L G C+EG L LVYEYMEN +L++ L +E++ + L+ TRMKI
Sbjct: 182 EIGMISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKI 361
Query: 248 ALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHV 307
+ +A+GL Y+H+ S +HRDIK+ N+LL+++ K++DFGL KL + N+ +T +
Sbjct: 362 CVGIAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHIST-RI 538
Query: 308 AGTFG 312
AGT G
Sbjct: 539 AGTIG 553
>TC18676 similar to UP|Q8RYS1 (Q8RYS1) Protein kinase-like, partial (15%)
Length = 814
Score = 148 bits (373), Expect = 2e-36
Identities = 87/214 (40%), Positives = 124/214 (57%), Gaps = 4/214 (1%)
Frame = +3
Query: 233 NSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLT 292
N ++ TR++IALD ARGLEYIH+H+ Y+H+DI + NILL+ +F K++DFGL
Sbjct: 18 NKGHSSLSWITRVQIALDAARGLEYIHEHTKTRYVHQDINTSNILLDASFRAKISDFGLA 197
Query: 293 KL-TDAANSADNTVHVAGTFGYMPPENAYGRI-SRKIDVYAFGVVLYELISAKAAVIKID 350
KL ++ T T+GY+ PE RI + K DVYAFGVVLYE+IS K A+I+
Sbjct: 198 KLVSETIEGGTTTTKGVSTYGYLAPEYLSNRIATSKSDVYAFGVVLYEIISGKKAIIQTQ 377
Query: 351 KTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCID--DLRKLVDPRLGYNYSIDSI 408
T+ E +SL ++ EV+ D + +R VDP + YS D +
Sbjct: 378 GTQG-------------PERRSLASIMLEVLRTVPDSLSTPSIRNHVDPIMKDLYSHDCV 518
Query: 409 SKMAKLAKACINRDPKQRPTMRDVVVSLMELNSS 442
+MA LAK C+ DP RP M+ VV+SL +++ S
Sbjct: 519 LQMAMLAKQCVEEDPILRPDMKQVVLSLSQIHLS 620
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 146 bits (368), Expect = 8e-36
Identities = 83/217 (38%), Positives = 135/217 (61%), Gaps = 12/217 (5%)
Frame = +1
Query: 112 TPSTKDVDKDT-----NDDNGSKYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVY 166
TP ++++ K + + N ++ P++SY+++ AT NF+ +GQG FG VY
Sbjct: 103 TPRSENLQKKSAFSWWSHQNNDQFASPSGIPKYSYKDIQKATQNFTTI--LGQGSFGTVY 276
Query: 167 YGEL-RGQKIAIKKM---KMQATREFLSELKVLTSVHHWNLVHLIGYCVE-GFLFLVYEY 221
+ G+ +A+K + Q EF +E+ +L +HH NLV+L+G+CV+ G LVY++
Sbjct: 277 KATMPTGEVVAVKVLAPNSKQGEHEFQTEVHLLGRLHHRNLVNLVGFCVDKGQRILVYQF 456
Query: 222 MENGNLSQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNEN 281
M NG+L+ L+ EKE ++ R++IA+D++ G+EY+H+ +VP IHRD+KS NILL+++
Sbjct: 457 MSNGSLANLLYGEEKE-LSWDERLQIAMDISHGIEYLHEGAVPPVIHRDLKSANILLDDS 633
Query: 282 FTGKVADFGLTK--LTDAANSADNTVHVAGTFGYMPP 316
VADFGL+K + D NS + GT+GYM P
Sbjct: 634 MRAMVADFGLSKEEIFDGRNSG-----LKGTYGYMDP 729
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 142 bits (359), Expect = 9e-35
Identities = 102/317 (32%), Positives = 162/317 (50%), Gaps = 13/317 (4%)
Frame = +1
Query: 133 VDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGEL-RGQKIAIKKMKM---QATREF 188
+D + F + +++AT++FSL+ K+G+GGFG VY G L GQ+IA+K++ Q EF
Sbjct: 1618 IDLATIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEEF 1797
Query: 189 LSELKVLTSVHHWNLVHLIGYCVEGFLFLVYEYMENGNLSQH----LHNSEKEPMTLSTR 244
+E+K++ + H NLV L G V EN + ++ L ++ + + + R
Sbjct: 1798 KNEIKLIARLQHRNLVKLFGCSVH--------QDENSHANKKMKILLDSTRSKLVDWNKR 1953
Query: 245 MKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNT 304
++I +ARGL Y+H S IHRD+K+ NILL++ K++DFGL ++ T
Sbjct: 1954 LQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEART 2133
Query: 305 VHVAGTFGYMPPENA-YGRISRKIDVYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKT 363
V GT+GYMPPE A +G S K DV++FGV++ E+IS K K
Sbjct: 2134 KRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGK------------------KI 2259
Query: 364 NESIDEYKSLVALFDE----VMNQTGDCIDDLRKLVDPRLGYNYSIDSISKMAKLAKACI 419
D + L L + + + +D+L L DP + I + +A C+
Sbjct: 2260 GRFYDPHHHLNLLSHAWRLWIEERPLELVDEL--LDDPVIP-----TEILRYIHVALLCV 2418
Query: 420 NRDPKQRPTMRDVVVSL 436
R P+ RP M +V+ L
Sbjct: 2419 QRRPENRPDMLSIVLML 2469
>TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3, complete
Length = 2481
Score = 135 bits (341), Expect = 1e-32
Identities = 105/352 (29%), Positives = 172/352 (48%), Gaps = 10/352 (2%)
Frame = +1
Query: 95 RKKNGEEESKFPPEDSMTPSTKDVDKDTNDDNGSKYIWVDKSPEFSYEELANATDNFSLA 154
R+K E E + E + KD D + +D + F + +++ T++FS +
Sbjct: 1312 RRKKNEREDEGGIETRIINHWKDKRGDED---------IDLATIFDFSTISSTTNHFSES 1464
Query: 155 KKIGQGGFGEVYYGEL-RGQKIAIKKMKM---QATREFLSELKVLTSVHHWNLVHLIGYC 210
K+G+GGFG VY G L GQ+IA+K++ Q EF +E+K++ + H NLV L+G
Sbjct: 1465 NKLGEGGFGPVYKGVLANGQEIAVKRLSNTSGQGMEEFKNEVKLIARLQHRNLVKLLGCS 1644
Query: 211 VE-GFLFLVYEYMENGNLSQHLHNSEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHR 269
+ + L+YE+M N +L + +S R++I IHR
Sbjct: 1645 IHHDEMLLIYEFMHNRSLDYFIFDS---------RLRI-------------------IHR 1740
Query: 270 DIKSDNILLNENFTGKVADFGLTKLTDAANSADNTVHVAGTFGYMPPENA-YGRISRKID 328
D+K+ NILL+ K++DFGL ++ T V GT+GYM PE A +G S K D
Sbjct: 1741 DLKTSNILLDSEMNPKISDFGLARIFTGDQVEAKTKRVMGTYGYMSPEYAVHGSFSVKSD 1920
Query: 329 VYAFGVVLYELISAKAAVIKIDKTEFEFKSLEIKTNESIDEYKSL-VALFDEVMNQTGDC 387
V++FGV++ E+IS K + + L +N ++ K+L + +F+ V N+
Sbjct: 1921 VFSFGVIVLEIISGK-KIGRFCDPHHHRNLLSHSSNFAVFLIKALRICMFENVKNRKAWR 2097
Query: 388 I---DDLRKLVDPRLGYNYSIDSISKMAKLAKACINRDPKQRPTMRDVVVSL 436
+ + +LVD L I + +A C+ + P+ RP M VV+ L
Sbjct: 2098 LWIEERPLELVDELLDGLAIPTEILRYIHIALLCVQQRPEYRPDMLSVVLML 2253
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 132 bits (333), Expect = 9e-32
Identities = 77/191 (40%), Positives = 110/191 (57%), Gaps = 11/191 (5%)
Frame = +3
Query: 138 EFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQKIAI-----KKMKMQATREFLSEL 192
EF+Y EL AT+NF K+GQGG+G VY G L +K+ + + KM++T +FLSEL
Sbjct: 129 EFNYVELKKATNNFDEKHKLGQGGYGVVYRGMLPKEKLEVAVKMFSRDKMKSTDDFLSEL 308
Query: 193 KVLTSVHHWNLVHLIGYCVE-GFLFLVYEYMENGNLSQHLHNSE----KEPMTLSTRMKI 247
++ + H +LV L G+C + G L LVY+YM NG+L H+ E P++ R KI
Sbjct: 309 IIINRLRHKHLVRLQGWCHKNGVLLLVYDYMPNGSLDSHIFCEEGGTITTPLSWPLRYKI 488
Query: 248 ALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTK-LTDAANSADNTVH 306
VA L Y+H+ +HRD+K+ NI+L+ F K+ DFGL + L + S
Sbjct: 489 ISGVASALNYLHNEYDQKVVHRDLKASNIMLDSEFNAKLGDFGLARALENEKISYTELEG 668
Query: 307 VAGTFGYMPPE 317
V GT GY+ PE
Sbjct: 669 VHGTMGYIAPE 701
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 132 bits (331), Expect = 2e-31
Identities = 76/172 (44%), Positives = 106/172 (61%), Gaps = 6/172 (3%)
Frame = +3
Query: 139 FSYEELANATDNFSLAKKIGQGGFGEVYYGELR-GQKIAIKKMKMQATR---EFLSELKV 194
FSYE L AT NF K+G+GGFG V+ G+L G++IA+KK+ ++ + +F++E K+
Sbjct: 207 FSYETLVAATKNFHAVNKLGEGGFGPVFKGKLNDGREIAVKKLSRRSNQGRTQFINEAKL 386
Query: 195 LTSVHHWNLVHLIGYCVEGF-LFLVYEYMENGNLSQHLHNSEK-EPMTLSTRMKIALDVA 252
LT V H N+V L GYC G LVYEY+ +L + L S+K E + R I VA
Sbjct: 387 LTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLLFRSQKKEQLDWKRRFDIISGVA 566
Query: 253 RGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTKLTDAANSADNT 304
RGL Y+H+ S IHRDIK+ NILL+E + K+ADFGL ++ + NT
Sbjct: 567 RGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADFGLARIFPEDQTHVNT 722
>CB829103
Length = 547
Score = 130 bits (327), Expect = 4e-31
Identities = 68/104 (65%), Positives = 84/104 (80%)
Frame = +1
Query: 345 AVIKIDKTEFEFKSLEIKTNESIDEYKSLVALFDEVMNQTGDCIDDLRKLVDPRLGYNYS 404
AV++I ++ E KSLEIKT+E E+KSLVALFDEV++ G+ I+ LRKLVDPRLG NYS
Sbjct: 1 AVVEIKESSTELKSLEIKTDEPSVEFKSLVALFDEVIDHEGNPIEGLRKLVDPRLGENYS 180
Query: 405 IDSISKMAKLAKACINRDPKQRPTMRDVVVSLMELNSSIDNKNS 448
IDSI +MA+LAKAC +RDPKQRP MR VVV LM LNS+ D++ S
Sbjct: 181 IDSIREMAQLAKACTDRDPKQRPPMRSVVVVLMALNSATDDRMS 312
>TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protein, partial
(42%)
Length = 946
Score = 122 bits (307), Expect = 9e-29
Identities = 72/180 (40%), Positives = 106/180 (58%), Gaps = 15/180 (8%)
Frame = +3
Query: 129 KYIWVDKSPEFSYEELANATDNFSLAKKIGQGGFGEVYYGELRGQK-----------IAI 177
+++ V K FS+ +L +AT +F IG+GGFG+VY G L +K +A+
Sbjct: 390 EFLEVAKLKVFSFGDLKSATKSFKADALIGEGGFGKVYKGWLDEKKLSPTKPGSGIMVAV 569
Query: 178 KKMK---MQATREFLSELKVLTSVHHWNLVHLIGYCVEGFLFL-VYEYMENGNLSQHLHN 233
KK+ MQ E+ SE+ L + H NLV L+GYC + FL VYE+M G+L HL
Sbjct: 570 KKLNPESMQGFHEWQSEINFLGRISHPNLVKLLGYCRDDEEFLLVYEFMPRGSLENHLFR 749
Query: 234 SEKEPMTLSTRMKIALDVARGLEYIHDHSVPVYIHRDIKSDNILLNENFTGKVADFGLTK 293
E ++ +TR+KIA+ ARGL ++H + I+RD K+ NILL+ N+ K++ FGLTK
Sbjct: 750 RNTESLSWNTRLKIAIGAARGLAFLHS-LXKIVIYRDFKASNILLDGNYNAKISXFGLTK 926
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.140 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,500,122
Number of Sequences: 28460
Number of extensions: 82344
Number of successful extensions: 957
Number of sequences better than 10.0: 310
Number of HSP's better than 10.0 without gapping: 709
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 740
length of query: 451
length of database: 4,897,600
effective HSP length: 93
effective length of query: 358
effective length of database: 2,250,820
effective search space: 805793560
effective search space used: 805793560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC147960.1


