

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.5 - phase: 0
(459 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
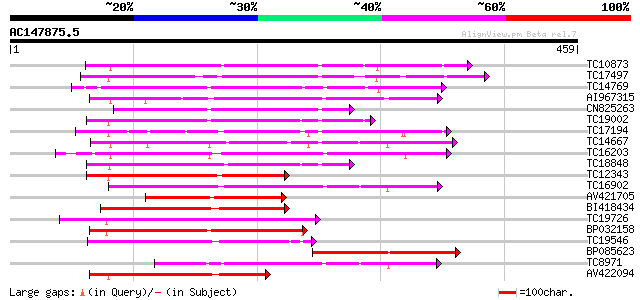
Score E
Sequences producing significant alignments: (bits) Value
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 174 2e-44
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 163 5e-41
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 162 8e-41
AI967315 156 8e-39
CN825263 154 3e-38
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 149 1e-36
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 148 2e-36
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 146 6e-36
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 144 2e-35
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 144 4e-35
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 143 7e-35
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 143 7e-35
AV421705 140 3e-34
BI418434 137 4e-33
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 135 2e-32
BP032158 133 5e-32
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 132 1e-31
BP085623 125 1e-29
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 122 1e-28
AV422094 121 3e-28
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 174 bits (442), Expect = 2e-44
Identities = 104/319 (32%), Positives = 172/319 (53%), Gaps = 6/319 (1%)
Frame = +1
Query: 62 ARRYSYAEVKRITNSFRDK--LGHGGYGVVYKASLTDGRQVAVKVINESKGNG-EEFINE 118
A + + E+ T +F++ +G GG+G VYK LT G VAVK ++ G +EF+ E
Sbjct: 406 AASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVME 585
Query: 119 VASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQ 178
V +S H N+V L+GYC + ++R L+YE+MP GSL+ +++ + +T +
Sbjct: 586 VLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNW--STRMK 759
Query: 179 IAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPG 238
+A+G ARGLEYLH +++ D+K NILLD F PK+SDFGLAK+ + D+
Sbjct: 760 VAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTR 939
Query: 239 TRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDW-- 296
GT GY APE G ++ KSD+YS+G+++LE++ GR+ T E W
Sbjct: 940 VMGTYGYCAPEY--AMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRR-PGEQNLVSWAR 1110
Query: 297 -IYKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSS 355
+ D + +++ L + + + ++ C+Q P RP + ++ L+ L+S
Sbjct: 1111PYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE-YLAS 1287
Query: 356 VTFPPKPVLFSPKRPPLQL 374
P L+ ++PP ++
Sbjct: 1288QGSPEAHYLYGVQQPPSRM 1344
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 163 bits (413), Expect = 5e-41
Identities = 119/347 (34%), Positives = 172/347 (49%), Gaps = 16/347 (4%)
Frame = +1
Query: 58 NLSIARRYSYAEVKRITNSFR--DKLGHGGYGVVYKASLTDGRQVAVKVINESKGNG-EE 114
++ +A + ++ + TN F +KLG GG+G VYK L +G+++AVK ++ + G G EE
Sbjct: 1615 DIDLATIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEE 1794
Query: 115 FINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSN 174
F NE+ I+R H N+V L G ++ + + M K + S V D N
Sbjct: 1795 FKNEIKLIARLQHRNLVKLFGCSVHQDENSHANKKM------KILLDSTRSKLV---DWN 1947
Query: 175 TLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIV 234
QI GIARGL YLHQ RI+H D+K NILLD+ PKISDFGLA+I +
Sbjct: 1948 KRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEA 2127
Query: 235 SIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFP 294
GT GYM PE G S KSDV+S+G+++LE+I G+K Y P
Sbjct: 2128 RTKRVMGTYGYMPPEY--AVHGSFSIKSDVFSFGVIVLEIISGKK--------IGRFYDP 2277
Query: 295 D------------WI-YKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPP 341
WI + LE ++LL+ I E + + V+L C+Q P +RP
Sbjct: 2278 HHHLNLLSHAWRLWIEERPLELVDELLDDPVIPTE----ILRYIHVALLCVQRRPENRPD 2445
Query: 342 MNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQLSNMSSSDWQETNSI 388
M ++ ML G + P P ++ K P+ L + S T S+
Sbjct: 2446 MLSIVLMLNGE-KELPKPRLPAFYTGKHDPIWLGSPSRCSTSITISL 2583
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 162 bits (411), Expect = 8e-41
Identities = 108/307 (35%), Positives = 168/307 (54%), Gaps = 4/307 (1%)
Frame = +3
Query: 51 EVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKG 110
+ F++S ++ A Y EV T ++ +G GG+G VY+ +L DG++VAVKV + +
Sbjct: 2100 DFFIKSVSIQ-AFTLEYIEVA--TERYKTLIGEGGFGSVYRGTLNDGQEVAVKVRSSTST 2270
Query: 111 NG-EEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVC 169
G EF NE+ +S H N+V LLGYC E++++ L+Y FM GSL +Y G P
Sbjct: 2271 QGTREFDNELNLLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLY--GEPAKRK 2444
Query: 170 DCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKIC-Q 228
D T IA+G ARGL YLH ++H DIK NILLD + C K++DFG +K Q
Sbjct: 2445 ILDWPTRLSIALGAARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQ 2624
Query: 229 MNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCT 288
DS VS+ RGT GY+ PE + +S KSDV+S+G+++LE++ GR+ T
Sbjct: 2625 EGDSYVSLE-VRGTAGYLDPEYYKTQ--QLSEKSDVFSFGVVLLEIVSGREPLNIKRPRT 2795
Query: 289 SEMYFPDWI--YKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVI 346
E +W Y + +++++ + + ++ V+L C++ RP M ++
Sbjct: 2796 -EWSLVEWATPYIRGSKVDEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIV 2972
Query: 347 EMLQGPL 353
L+ L
Sbjct: 2973 RELEDAL 2993
>AI967315
Length = 1308
Score = 156 bits (394), Expect = 8e-39
Identities = 98/291 (33%), Positives = 155/291 (52%), Gaps = 5/291 (1%)
Frame = +1
Query: 65 YSYAEVKRITNSFRDK--LGHGGYGVVYKASLTDGRQVAVKVINES---KGNGEEFINEV 119
+SY E+ TN F + +G GGY VYK L G ++AVK + + + +EF+ E+
Sbjct: 112 FSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEI 291
Query: 120 ASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQI 179
+I H N++ LLG C + N L++E GS+ I+ + + D T ++I
Sbjct: 292 GTIGHVCHSNVMPLLGCCID-NGLYLVFELSTVGSVASLIHD----EKMAPLDWKTRYKI 456
Query: 180 AIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGT 239
+G ARGL YLH+GC RI+H DIK NILL E+F P+ISDFGLAK + SI
Sbjct: 457 VLGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPI 636
Query: 240 RGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYK 299
GT G++APE + G V K+DV+++G+ +LE+I GRK P
Sbjct: 637 EGTFGHLAPEYYMH--GVVDEKTDVFAFGVFLLEVISGRKPVDGSHQSLHTWAKPILSKW 810
Query: 300 DLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQ 350
++E+ L++ + ++ + CI+ + RP M++V+E+++
Sbjct: 811 EIEK---LVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVME 954
>CN825263
Length = 663
Score = 154 bits (389), Expect = 3e-38
Identities = 84/196 (42%), Positives = 117/196 (58%), Gaps = 1/196 (0%)
Frame = +1
Query: 85 GYGVVYKASLTDGRQVAVKVIN-ESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKR 143
G+G+VYK L DGR VAVK++ + + G EF+ EV +SR H N+V L+G C E R
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTR 180
Query: 144 ALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDI 203
LIYE +P GS++ ++ G D N +IA+G ARGL YLH+ + ++H D
Sbjct: 181 CLIYELVPNGSVESHLH--GADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDF 354
Query: 204 KPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSD 263
K NILL+ +F PK+SDFGLA+ + GT GY+APE G + KSD
Sbjct: 355 KSSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEY--AMTGHLLVKSD 528
Query: 264 VYSYGMLILEMIGGRK 279
VYSYG+++LE++ G K
Sbjct: 529 VYSYGVVLLELLTGTK 576
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 149 bits (375), Expect = 1e-36
Identities = 90/237 (37%), Positives = 133/237 (55%), Gaps = 3/237 (1%)
Frame = +1
Query: 63 RRYSYAEVKRITNSFR--DKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEV 119
R +S E+ TN+F +KLG GG+G VY L DG Q+AVK + + EF EV
Sbjct: 22 RVFSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEV 201
Query: 120 ASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQI 179
++R H N++SL GYC E +R ++Y++MP SL ++ G + C D N I
Sbjct: 202 EILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLH--GQHSSECLLDWNRRMNI 375
Query: 180 AIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGT 239
AIG A G+ YLH + I+H DIK N+LLD +F +++DFG AK+ + V+
Sbjct: 376 AIGSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVT-TRV 552
Query: 240 RGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDW 296
+GT+GY+APE G + DV+S+G+L+LE+ G+K + S T + DW
Sbjct: 553 KGTLGYLAPEY--AMLGKANECCDVFSFGILLLELASGKKPLEKLSS-TVKRSINDW 714
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 148 bits (374), Expect = 2e-36
Identities = 102/315 (32%), Positives = 172/315 (54%), Gaps = 11/315 (3%)
Frame = +1
Query: 54 MQSYNLSIARRYSYAEVKRITNSFR--DKLGHGGYGVVYKASLTDGRQVAVKVINESKGN 111
+ S ++ + +SY E+ + TN+F +K+G GG+G VY A L G++ A+K ++
Sbjct: 1018 LTSIMVAKSMEFSYQELAKATNNFSLDNKIGQGGFGAVYYAELR-GKKTAIKKMDVQAST 1194
Query: 112 GEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDC 171
EF+ E+ ++ H+N+V L+GYC E + L+YE + G+L ++++ SG
Sbjct: 1195 --EFLCELKVLTHVHHLNLVRLIGYCVEGSL-FLVYEHIDNGNLGQYLHGSGKEPLPW-- 1359
Query: 172 DSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMND 231
++ QIA+ ARGLEY+H+ +H D+K NIL+D+N K++DFGL K+ ++ +
Sbjct: 1360 --SSRVQIALDAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGN 1533
Query: 232 SIVSIPGTR--GTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTS 289
S + TR GT GYM PE +G +S K DVY++G+++ E+I + G +
Sbjct: 1534 STLQ---TRLVGTFGYMPPEY--AQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVA 1698
Query: 290 EMYFPDWIYKDLEQGNDLLNSLTISEE----EN---DMVKKITMVSLWCIQTNPLDRPPM 342
E ++++ +D ++L + EN D V KI + C + NPL RP M
Sbjct: 1699 ESKGLVALFEEALNKSDPCDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSM 1878
Query: 343 NKVIEMLQGPLSSVT 357
++ L LSS+T
Sbjct: 1879 RSLVVALM-TLSSLT 1920
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 146 bits (369), Expect = 6e-36
Identities = 107/310 (34%), Positives = 153/310 (48%), Gaps = 13/310 (4%)
Frame = +2
Query: 66 SYAEVKRITNSFRDK--LGHGGYGVVYKASLTDGRQVAVKVINESKG--NGEEFINEVAS 121
S E+K T++F K +G G YG VY A+L DG VAVK ++ S EF+ +V+
Sbjct: 326 SLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNEFLTQVSM 505
Query: 122 ISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY-----KSGFPDAVCDCDSNTL 176
+SR + N V L GYC E N R L YEF GSL ++ + P D
Sbjct: 506 VSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRV- 682
Query: 177 FQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSI 236
+IA+ ARGLEYLH+ I+H DI+ N+L+ E++ KI+DF L+ Q D +
Sbjct: 683 -RIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSN--QAPDMAARL 853
Query: 237 PGTR--GTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFP 294
TR GT GY APE G ++ KSDVYS+G+++LE++ GRK +
Sbjct: 854 HSTRVLGTFGYHAPEY--AMTGQLTQKSDVYSFGVVLLELLTGRKPVD-HTMPRGQQSLV 1024
Query: 295 DWIYKDLEQG--NDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGP 352
W L + ++ E V K+ V+ C+Q RP M+ V++ LQ
Sbjct: 1025TWATPRLSEDKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQPL 1204
Query: 353 LSSVTFPPKP 362
L + P P
Sbjct: 1205LKTPAAAPAP 1234
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 144 bits (364), Expect = 2e-35
Identities = 103/331 (31%), Positives = 168/331 (50%), Gaps = 11/331 (3%)
Frame = +2
Query: 38 IFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDK--LGHGGYGVVYKASLT 95
+ RKRR H Q++ L+ +R + + + +++ +G GG G+VY+ S+
Sbjct: 2099 VVRKRRLHR-------AQAWKLTAFQRLEI-KAEDVVECLKEENIIGKGGAGIVYRGSMP 2254
Query: 96 DGRQVAVK-VINESKG-NGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKG 153
+G VA+K ++ + G N F E+ ++ + H NI+ LLGY + L+YE+MP G
Sbjct: 2255 NGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNG 2434
Query: 154 SLDKFIY--KSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLD 211
SL ++++ K G ++IA+ ARGL Y+H CS I+H D+K NILLD
Sbjct: 2435 SLGEWLHGAKGGH------LRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLD 2596
Query: 212 ENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLI 271
+F ++DFGLAK + S+ G+ GY+APE V KSDVYS+G+++
Sbjct: 2597 ADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTL--KVDEKSDVYSFGVVL 2770
Query: 272 LEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEEEND-----MVKKITM 326
LE+I GRK G + + + +L Q +D L + + V +
Sbjct: 2771 LELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIHMFN 2950
Query: 327 VSLWCIQTNPLDRPPMNKVIEMLQGPLSSVT 357
+++ C++ RP M +V+ ML P S T
Sbjct: 2951 IAMMCVKEMGPARPTMREVVHMLTNPPQSNT 3043
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 144 bits (362), Expect = 4e-35
Identities = 84/220 (38%), Positives = 125/220 (56%), Gaps = 3/220 (1%)
Frame = +2
Query: 63 RRYSYAEVKRITNSFRD--KLGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEV 119
R ++Y E+ T F D KLG GG+G VY +DG Q+AVK + E EF EV
Sbjct: 260 RIFTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEV 439
Query: 120 ASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQI 179
+ R H N++ L GYC ++R ++Y++MP SL ++ + + +I
Sbjct: 440 EVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKR--MKI 613
Query: 180 AIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGT 239
AIG A G+ YLH + I+H DIK N+LL+ +F P ++DFG AK+ S ++
Sbjct: 614 AIGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMT-TRV 790
Query: 240 RGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRK 279
+GT+GY+APE +G VS DVYS+G+L+LE++ GRK
Sbjct: 791 KGTLGYLAPEY--AMWGKVSESCDVYSFGILLLELVTGRK 904
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 143 bits (360), Expect = 7e-35
Identities = 73/167 (43%), Positives = 106/167 (62%), Gaps = 3/167 (1%)
Frame = +3
Query: 63 RRYSYAEVKRITNSFR--DKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEV 119
+ +SY + T +F +KLG GG+G V+K L DGR++AVK ++ G +FINE
Sbjct: 201 KTFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLNDGREIAVKKLSRRSNQGRTQFINEA 380
Query: 120 ASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQI 179
++R H N+VSL GYC +++ L+YE++P+ SLDK +++S + + D F I
Sbjct: 381 KLLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLLFRSQKKEQL---DWKRRFDI 551
Query: 180 AIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKI 226
G+ARGL YLH+ I+H DIK NILLDE + PKI+DFGLA+I
Sbjct: 552 ISGVARGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADFGLARI 692
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 143 bits (360), Expect = 7e-35
Identities = 89/273 (32%), Positives = 140/273 (50%), Gaps = 3/273 (1%)
Frame = +3
Query: 81 LGHGGYGVVYKASLTDGRQVAVKVINESKGNG-EEFINEVASISRTSHMNIVSLLGYCYE 139
LG GG+G VY + G +VA+K N G EF E+ +S+ H ++VSL+GYC E
Sbjct: 15 LGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYCEE 194
Query: 140 ANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRIL 199
+ L+Y+ M G+L + +YK+ P +I IG ARGL YLH G I+
Sbjct: 195 NTEMILVYDHMAYGTLREHLYKTQKPPLPW----KQRLEICIGAARGLHYLHTGAKYTII 362
Query: 200 HLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVS 259
H D+K NILLDE + K+SDFGL+K D+ +G+ GY+ PE F R ++
Sbjct: 363 HRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQ--QLT 536
Query: 260 YKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQG--NDLLNSLTISEEE 317
KSDVYS+G+++ E++ R ++ +W +G + +L+ +
Sbjct: 537 DKSDVYSFGVVLFEILCARPALNP-SLAKEQVSLAEWASHCYNKGILDQILDPYLKGKIA 713
Query: 318 NDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQ 350
+ KK ++ C+ ++RP M V+ L+
Sbjct: 714 PECFKKFAETAMKCVSDQGIERPSMGDVLWNLE 812
>AV421705
Length = 341
Score = 140 bits (354), Expect = 3e-34
Identities = 64/114 (56%), Positives = 87/114 (76%)
Frame = +3
Query: 111 NGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCD 170
NG++FI+EVA+I R H+N+V L+GYC + KRAL+YEFMP GSLDK+I+ +
Sbjct: 9 NGQDFISEVATIGRIHHINVVRLVGYCVDGKKRALVYEFMPNGSLDKYIFSK---EGSAW 179
Query: 171 CDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLA 224
+ +++I++GIARG+ YLHQGC +ILH DIKP NILLD++F PK+SDFGLA
Sbjct: 180 LSYDQIYEISLGIARGVAYLHQGCDMQILHFDIKPHNILLDKDFIPKVSDFGLA 341
>BI418434
Length = 478
Score = 137 bits (345), Expect = 4e-33
Identities = 70/154 (45%), Positives = 98/154 (63%), Gaps = 1/154 (0%)
Frame = +2
Query: 74 TNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTSHMNIVSL 133
T F +K+GHGG+G V++ L+D VAVK + G +EF EV++I H+N+V L
Sbjct: 14 TRGFSEKVGHGGFGTVFQGELSDSSLVAVKRLERPGGGEKEFRAEVSTIGNIQHVNLVRL 193
Query: 134 LGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDS-NTLFQIAIGIARGLEYLHQ 192
G+C E + R L+YE+M G+L ++ K G C S + FQ+AIG A+G+ YLH+
Sbjct: 194 RGFCSENSHRLLVYEYMHNGALSAYLRKDG------PCLSWDVRFQVAIGTAKGIAYLHE 355
Query: 193 GCSSRILHLDIKPQNILLDENFCPKISDFGLAKI 226
ILH DIKP+NILLD +F K+SDFGLAK+
Sbjct: 356 ELRCCILHCDIKPENILLDSDFTAKVSDFGLAKL 457
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 135 bits (339), Expect = 2e-32
Identities = 75/217 (34%), Positives = 122/217 (55%), Gaps = 6/217 (2%)
Frame = +3
Query: 41 KRRKHVDNNVEVFMQSYNL-SIARRYSYAEVKRITNSF--RDKLGHGGYGVVYKASLTDG 97
K+++ +N ++ +L R ++Y E+K+ TN+F + KLG GGYGVVY+ L
Sbjct: 57 KKKREEGSNSQILGTLKSLPGTPREFNYVELKKATNNFDEKHKLGQGGYGVVYRGMLPKE 236
Query: 98 R-QVAVKVINESKGNG-EEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSL 155
+ +VAVK+ + K ++F++E+ I+R H ++V L G+C++ L+Y++MP GSL
Sbjct: 237 KLEVAVKMFSRDKMKSTDDFLSELIIINRLRHKHLVRLQGWCHKNGVLLLVYDYMPNGSL 416
Query: 156 DKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFC 215
D I+ ++I G+A L YLH +++H D+K NI+LD F
Sbjct: 417 DSHIFCEEGGTITTPLSWPLRYKIISGVASALNYLHNEYDQKVVHRDLKASNIMLDSEFN 596
Query: 216 PKISDFGLAKICQMND-SIVSIPGTRGTIGYMAPEVF 251
K+ DFGLA+ + S + G GT+GY+APE F
Sbjct: 597 AKLGDFGLARALENEKISYTELEGVHGTMGYIAPECF 707
>BP032158
Length = 555
Score = 133 bits (335), Expect = 5e-32
Identities = 75/182 (41%), Positives = 113/182 (61%), Gaps = 5/182 (2%)
Frame = +2
Query: 65 YSYAEVKRITNSF--RDKLGHGGYGVVYKASLTDGRQVAVKVIN-ESKGNGEEFINEVAS 121
+S ++K TN+F +K+G GG+G VYK L++G +AVK ++ +SK EFINE+
Sbjct: 14 FSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSSKSKQGNREFINEIGM 193
Query: 122 ISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAI 181
IS H N+V L G C E N+ L+YE+M SL + ++ G + + + T +I +
Sbjct: 194 ISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALF--GNEEQKLNLNWRTRMKICV 367
Query: 182 GIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVS--IPGT 239
GIA+GL YLH+ +I+H DIK N+LLD++ KISDFGLAK+ + ++ +S I GT
Sbjct: 368 GIAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHISTRIAGT 547
Query: 240 RG 241
G
Sbjct: 548 IG 553
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 132 bits (332), Expect = 1e-31
Identities = 73/186 (39%), Positives = 111/186 (59%), Gaps = 1/186 (0%)
Frame = +1
Query: 64 RYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEVASI 122
+YSY ++++ T +F LG G +G VYKA++ G VAVKV+ + GE EF EV +
Sbjct: 196 KYSYKDIQKATQNFTTILGQGSFGTVYKATMPTGEVVAVKVLAPNSKQGEHEFQTEVHLL 375
Query: 123 SRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIG 182
R H N+V+L+G+C + +R L+Y+FM GSL +Y + + QIA+
Sbjct: 376 GRLHHRNLVNLVGFCVDKGQRILVYQFMSNGSLANLLYGEEK-----ELSWDERLQIAMD 540
Query: 183 IARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGT 242
I+ G+EYLH+G ++H D+K NILLD++ ++DFGL+K ++ D S G +GT
Sbjct: 541 ISHGIEYLHEGAVPPVIHRDLKSANILLDDSMRAMVADFGLSKE-EIFDGRNS--GLKGT 711
Query: 243 IGYMAP 248
GYM P
Sbjct: 712 YGYMDP 729
>BP085623
Length = 528
Score = 125 bits (314), Expect = 1e-29
Identities = 60/120 (50%), Positives = 81/120 (67%)
Frame = -3
Query: 246 MAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQGN 305
MAPE+F + GGVSYK DVYS+GML++EM RKN +S++YFP WIY L + N
Sbjct: 526 MAPELFYKNIGGVSYKXDVYSFGMLLMEMASNRKNLNPHVEHSSQLYFPFWIYDQLGKQN 347
Query: 306 DLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLF 365
D + + EEE + KK+ MV+LWCIQ P DRP M +V+EML+G + ++ PPKP L+
Sbjct: 346 D-IELEDVKEEEVKIAKKMIMVALWCIQLKPDDRPSMKEVVEMLEGDVENLEMPPKPSLY 170
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 122 bits (306), Expect = 1e-28
Identities = 78/235 (33%), Positives = 125/235 (53%), Gaps = 3/235 (1%)
Frame = +3
Query: 118 EVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLF 177
EV +++ H N+V LLG+ + N+R LI E++P G+L + + G + D N
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHL--DGLRGKILDF--NQRL 173
Query: 178 QIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSI- 236
+IAI +A GL YLH +I+H D+K NILL E+ K++DFG A++ +N I
Sbjct: 174 EIAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHIS 353
Query: 237 PGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDW 296
+GT+GY+ PE ++ KSDVYS+G+L+LE++ GR+ + E W
Sbjct: 354 TKVKGTVGYLDPEYMKT--HQLTPKSDVYSFGILLLEILTGRRPVEL-KKAADERVTLRW 524
Query: 297 IYKDLEQGN--DLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEML 349
++ +G+ +L++ L D++ K+ +S C DRP M V E L
Sbjct: 525 AFRKYNEGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSVGEQL 689
>AV422094
Length = 462
Score = 121 bits (303), Expect = 3e-28
Identities = 64/150 (42%), Positives = 92/150 (60%), Gaps = 3/150 (2%)
Frame = +2
Query: 65 YSYAEVKRITNSFR--DKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEVAS 121
+SY+E+K TN F +KLG GG+G VYK L DG +AVK ++ G+ +FI E+A+
Sbjct: 14 FSYSELKNATNDFNIDNKLGEGGFGPVYKGILNDGTVIAVKQLSLGSHQGKSQFIAEIAT 193
Query: 122 ISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAI 181
IS H N+V L G C E +KR L+YE++ SLD+ ++ + +T + I +
Sbjct: 194 ISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQALFGKAL-----SLNWSTRYDICL 358
Query: 182 GIARGLEYLHQGCSSRILHLDIKPQNILLD 211
G+ARGL YLH+ RI+H D+K NILLD
Sbjct: 359 GVARGLTYLHEESRLRIVHRDVKASNILLD 448
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,145,849
Number of Sequences: 28460
Number of extensions: 99784
Number of successful extensions: 1062
Number of sequences better than 10.0: 298
Number of HSP's better than 10.0 without gapping: 860
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 874
length of query: 459
length of database: 4,897,600
effective HSP length: 93
effective length of query: 366
effective length of database: 2,250,820
effective search space: 823800120
effective search space used: 823800120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC147875.5


