

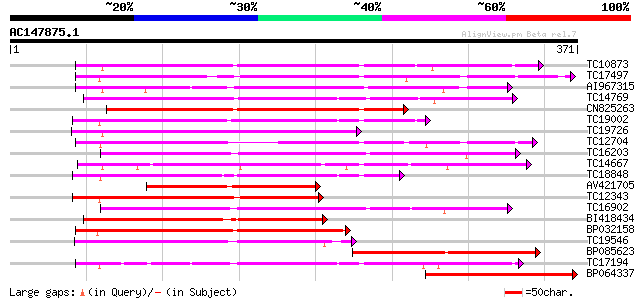
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.1 + phase: 0
(371 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 172 6e-44
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 165 1e-41
AI967315 165 1e-41
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 164 3e-41
CN825263 157 3e-39
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 151 1e-37
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 150 3e-37
TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3,... 148 1e-36
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 147 3e-36
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 147 4e-36
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 146 6e-36
AV421705 143 5e-35
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 142 1e-34
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 140 3e-34
BI418434 139 8e-34
BP032158 138 1e-33
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 137 2e-33
BP085623 127 4e-30
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 127 4e-30
BP064337 125 1e-29
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 172 bits (437), Expect = 6e-44
Identities = 102/312 (32%), Positives = 168/312 (53%), Gaps = 6/312 (1%)
Frame = +1
Query: 44 YSYAEVKMITNYFREK--LGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFINEVAS 100
+ + E+ T F+E +G+GG+G VYK L VAVK +S G +EF+ EV
Sbjct: 415 FGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLM 594
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
+S H N+V L+GYC + ++R L+YE+MP GSL+ +++ +W+T ++A+
Sbjct: 595 LSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKE--PLNWSTRMKVAV 768
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
G A+GLEYLH +++ D+K NILLD +F PK+SDFGLAK+ D+ G
Sbjct: 769 GAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMG 948
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDW---IY 277
T GY APE G ++ +SD+YS+G+++LE++ GR+ DT E W +
Sbjct: 949 TYGYCAPEY--AMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRR-PGEQNLVSWARPYF 1119
Query: 278 KDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSY 337
D + +++ + + + ++ C+Q +P RP + ++ L+ L+S
Sbjct: 1120SDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE-YLASQGS 1296
Query: 338 PPKPVLYSPERP 349
P LY ++P
Sbjct: 1297PEAHYLYGVQQP 1332
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 165 bits (417), Expect = 1e-41
Identities = 117/335 (34%), Positives = 173/335 (50%), Gaps = 8/335 (2%)
Frame = +1
Query: 44 YSYAEVKMITNYFR--EKLGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFINEVAS 100
+ ++ + TN+F KLG+GG+G VYK L N +++AVK +S T G G EEF NE+
Sbjct: 1636 FDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEEFKNEIKL 1815
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
I+R H N+V L G +++ + + M K + S + DWN +I
Sbjct: 1816 IARLQHRNLVKLFGCSVHQDENSHANKKM------KILLDSTRSKLV---DWNKRLQIID 1968
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
GIA+GL YLHQ RI+H D+K NILLD++ PKISDFGLA+I G
Sbjct: 1969 GIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMG 2148
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRK---NYDTGGSCTSEMY-FPDWI 276
T GYM PE G S +SDV+S+G+++LE+I G+K YD + + WI
Sbjct: 2149 TYGYMPPEY--AVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRLWI 2322
Query: 277 -YKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSV 335
+ LE + LL+ I E I + V+L C+Q +P +RP M ++ ML G +
Sbjct: 2323 EERPLELVDELLDDPVIPTE----ILRYIHVALLCVQRRPENRPDMLSIVLMLNGE-KEL 2487
Query: 336 SYPPKPVLYSPERPELQVSDMSSSDLYETNSVTVS 370
P P Y+ + + + S + S+T+S
Sbjct: 2488 PKPRLPAFYTGKHDPIWLGSPSRC----STSITIS 2580
>AI967315
Length = 1308
Score = 165 bits (417), Expect = 1e-41
Identities = 100/293 (34%), Positives = 161/293 (54%), Gaps = 7/293 (2%)
Frame = +1
Query: 44 YSYAEVKMITNYFREK--LGQGGYGVVYKASLYNSRQVAVKVISET---KGNGEEFINEV 98
+SY E+ TN F + +G+GGY VYK L + ++AVK ++ T + +EF+ E+
Sbjct: 112 FSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEI 291
Query: 99 ASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRI 158
+I H N++ LLG C + N L++E GS+ I+ + + DW T ++I
Sbjct: 292 GTIGHVCHSNVMPLLGCCID-NGLYLVFELSTVGSVASLIHDEK----MAPLDWKTRYKI 456
Query: 159 AIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGA 218
+G A+GL YLH+GC RI+H DIK NILL EDF P+ISDFGLAK + + SI
Sbjct: 457 VLGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPI 636
Query: 219 RGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYK 278
GT G++APE + G V ++DV+++G+ +LE+I GRK D P
Sbjct: 637 EGTFGHLAPEYYMH--GVVDEKTDVFAFGVFLLEVISGRKPVDGSHQSLHTWAKPILSKW 810
Query: 279 DLEQ--GNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQ 329
++E+ L C +++ ++ + CI+ + RP M++V+E+++
Sbjct: 811 EIEKLVDPRLEGCYDVTQ-----FNRVAFAASLCIRASSTWRPTMSEVLEVME 954
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 164 bits (414), Expect = 3e-41
Identities = 101/288 (35%), Positives = 159/288 (55%), Gaps = 4/288 (1%)
Frame = +3
Query: 49 VKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFINEVASISRTSHM 107
+++ T ++ +G+GG+G VY+ +L + ++VAVKV S T G EF NE+ +S H
Sbjct: 2148 IEVATERYKTLIGEGGFGSVYRGTLNDGQEVAVKVRSSTSTQGTREFDNELNLLSAIQHE 2327
Query: 108 NIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLE 167
N+V LLGYC E +++ L+Y FM GSL +Y I DW T IA+G A+GL
Sbjct: 2328 NLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKI--LDWPTRLSIALGAARGLA 2501
Query: 168 YLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKIC-QRKDSIVSILGARGTIGYMA 226
YLH ++H DIK NILLD C K++DFG +K Q DS VS L RGT GY+
Sbjct: 2502 YLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVS-LEVRGTAGYLD 2678
Query: 227 PEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWI--YKDLEQGN 284
PE + +S +SDV+S+G+++LE++ GR+ + T E +W Y + +
Sbjct: 2679 PEYYKTQ--QLSEKSDVFSFGVVLLEIVSGREPLNIKRPRT-EWSLVEWATPYIRGSKVD 2849
Query: 285 TLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPL 332
+++ + + ++ V+L C++ + RP M ++ L+ L
Sbjct: 2850 EIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVRELEDAL 2993
>CN825263
Length = 663
Score = 157 bits (396), Expect = 3e-39
Identities = 82/199 (41%), Positives = 121/199 (60%), Gaps = 1/199 (0%)
Frame = +1
Query: 64 GYGVVYKASLYNSRQVAVKVIS-ETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKR 122
G+G+VYK L + R VAVK++ + + G EF+ EV +SR H N+V L+G C E+ R
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTR 180
Query: 123 ALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDI 182
LIYE +P GS++ ++ ++ DWN +IA+G A+GL YLH+ + ++H D
Sbjct: 181 CLIYELVPNGSVESHLHGADKETG--PLDWNARMKIALGAARGLAYLHEDSNPCVIHRDF 354
Query: 183 KPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSD 242
K NILL+ DF PK+SDFGLA+ + + GT GY+APE G + +SD
Sbjct: 355 KSSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEY--AMTGHLLVKSD 528
Query: 243 VYSYGMLILEMIGGRKNYD 261
VYSYG+++LE++ G K D
Sbjct: 529 VYSYGVVLLELLTGTKPVD 585
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 151 bits (382), Expect = 1e-37
Identities = 89/237 (37%), Positives = 134/237 (55%), Gaps = 3/237 (1%)
Frame = +1
Query: 42 RRYSYAEVKMITNYFR--EKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEV 98
R +S E+ TN F KLG+GG+G VY L++ Q+AVK + + EF EV
Sbjct: 22 RVFSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEV 201
Query: 99 ASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRI 158
++R H N++SL GYC E +R ++Y++MP SL ++ ++ C DWN I
Sbjct: 202 EILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQH--SSECLLDWNRRMNI 375
Query: 159 AIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGA 218
AIG A+G+ YLH + I+H DIK N+LLD DF +++DFG AK+ + V+
Sbjct: 376 AIGSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVT-TRV 552
Query: 219 RGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDW 275
+GT+GY+APE G + DV+S+G+L+LE+ G+K + S T + DW
Sbjct: 553 KGTLGYLAPEY--AMLGKANECCDVFSFGILLLELASGKKPLEKLSS-TVKRSINDW 714
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 150 bits (380), Expect = 3e-37
Identities = 79/195 (40%), Positives = 116/195 (58%), Gaps = 5/195 (2%)
Frame = +3
Query: 41 PRRYSYAEVKMITNYFREK--LGQGGYGVVYKASLYNSR-QVAVKVISETKGNG-EEFIN 96
PR ++Y E+K TN F EK LGQGGYGVVY+ L + +VAVK+ S K ++F++
Sbjct: 123 PREFNYVELKKATNNFDEKHKLGQGGYGVVYRGMLPKEKLEVAVKMFSRDKMKSTDDFLS 302
Query: 97 EVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLF 156
E+ I+R H ++V L G+C++ L+Y++MP GSLD I+ E W +
Sbjct: 303 ELIIINRLRHKHLVRLQGWCHKNGVLLLVYDYMPNGSLDSHIFCEEGGTITTPLSWPLRY 482
Query: 157 RIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQ-RKDSIVSI 215
+I G+A L YLH +++H D+K NI+LD +F K+ DFGLA+ + K S +
Sbjct: 483 KIISGVASALNYLHNEYDQKVVHRDLKASNIMLDSEFNAKLGDFGLARALENEKISYTEL 662
Query: 216 LGARGTIGYMAPEIF 230
G GT+GY+APE F
Sbjct: 663 EGVHGTMGYIAPECF 707
>TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3, complete
Length = 2481
Score = 148 bits (374), Expect = 1e-36
Identities = 112/337 (33%), Positives = 154/337 (45%), Gaps = 35/337 (10%)
Frame = +1
Query: 44 YSYAEVKMITNYFRE--KLGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFINEVAS 100
+ ++ + TN+F E KLG+GG+G VYK L N +++AVK +S T G G EEF NEV
Sbjct: 1417 FDFSTISSTTNHFSESNKLGEGGFGPVYKGVLANGQEIAVKRLSNTSGQGMEEFKNEVKL 1596
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
I+R H N+V LLG ++ LIYEFM SLD FI+ S
Sbjct: 1597 IARLQHRNLVKLLGCSIHHDEMLLIYEFMHNRSLDYFIFDSRL----------------- 1725
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
RI+H D+K NILLD + PKISDFGLA+I G
Sbjct: 1726 ---------------RIIHRDLKTSNILLDSEMNPKISDFGLARIFTGDQVEAKTKRVMG 1860
Query: 221 TIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMY--------- 271
T GYM+PE G S +SDV+S+G+++LE+I G+K G C +
Sbjct: 1861 TYGYMSPEY--AVHGSFSVKSDVFSFGVIVLEIISGKK---IGRFCDPHHHRNLLSHSSN 2025
Query: 272 ----------------------FPDWI-YKDLEQGNTLLNCSTISEEENDMIRKITLVSL 308
+ WI + LE + LL+ I E I + ++L
Sbjct: 2026 FAVFLIKALRICMFENVKNRKAWRLWIEERPLELVDELLDGLAIPTE----ILRYIHIAL 2193
Query: 309 WCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYS 345
C+Q +P RP M V+ ML G + P P Y+
Sbjct: 2194 LCVQQRPEYRPDMLSVVLMLNGE-KELPKPSLPAFYT 2301
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 147 bits (371), Expect = 3e-36
Identities = 91/282 (32%), Positives = 152/282 (53%), Gaps = 7/282 (2%)
Frame = +2
Query: 60 LGQGGYGVVYKASLYNSRQVAVK-VISETKG-NGEEFINEVASISRTSHMNIVSLLGYCY 117
+G+GG G+VY+ S+ N VA+K ++ + G N F E+ ++ + H NI+ LLGY
Sbjct: 2210 IGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVS 2389
Query: 118 EENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRI 177
++ L+YE+MP GSL ++++ ++ + W ++IA+ A+GL Y+H CS I
Sbjct: 2390 NKDTNLLLYEYMPNGSLGEWLHGAKGGH----LRWEMRYKIAVEAARGLCYMHHDCSPLI 2557
Query: 178 LHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGV 237
+H D+K NILLD DF ++DFGLAK + S+ G+ GY+APE V
Sbjct: 2558 IHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTL--KV 2731
Query: 238 SYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDLEQ-GNTLLNCSTISEEE 296
+SDVYS+G+++LE+I GRK G + + + +L Q +T L + +
Sbjct: 2732 DEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRL 2911
Query: 297 N----DMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSS 334
+ + + +++ C++ RP M +V+ ML P S
Sbjct: 2912 SGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHMLTNPPQS 3037
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 147 bits (370), Expect = 4e-36
Identities = 107/309 (34%), Positives = 156/309 (49%), Gaps = 12/309 (3%)
Frame = +2
Query: 45 SYAEVKMITNYFREK--LGQGGYGVVYKASLYNSRQVAVK---VISETKGNGEEFINEVA 99
S E+K T+ F K +G+G YG VY A+L + VAVK V SE + N E F+ +V+
Sbjct: 326 SLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNE-FLTQVS 502
Query: 100 SISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICD---FDWNTLF 156
+SR + N V L GYC E N R L YEF GSL ++ + DW
Sbjct: 503 MVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRV 682
Query: 157 RIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSIL 216
RIA+ A+GLEYLH+ I+H DI+ N+L+ ED+ KI+DF L+ Q D +
Sbjct: 683 RIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSN--QAPDMAARLH 856
Query: 217 GAR--GTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPD 274
R GT GY APE G ++ +SDVYS+G+++LE++ GRK D +
Sbjct: 857 STRVLGTFGYHAPEY--AMTGQLTQKSDVYSFGVVLLELLTGRKPVD-HTMPRGQQSLVT 1027
Query: 275 WIYKDLEQGNT--LLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPL 332
W L + ++ E + K+ V+ C+Q + RP M+ V++ LQ L
Sbjct: 1028WATPRLSEDKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQPLL 1207
Query: 333 SSVSYPPKP 341
+ + P P
Sbjct: 1208KTPAAAPAP 1234
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 146 bits (368), Expect = 6e-36
Identities = 84/220 (38%), Positives = 129/220 (58%), Gaps = 3/220 (1%)
Frame = +2
Query: 42 RRYSYAEVKMITNYFRE--KLGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEV 98
R ++Y E+ T F + KLG+GG+G VY + Q+AVK + E EF EV
Sbjct: 260 RIFTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEV 439
Query: 99 ASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRI 158
+ R H N++ L GYC +++R ++Y++MP SL ++ +F + +W +I
Sbjct: 440 EVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLH-GQFAVEV-QLNWQKRMKI 613
Query: 159 AIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGA 218
AIG A+G+ YLH + I+H DIK N+LL+ DF P ++DFG AK+ S ++
Sbjct: 614 AIGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMT-TRV 790
Query: 219 RGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRK 258
+GT+GY+APE +G VS DVYS+G+L+LE++ GRK
Sbjct: 791 KGTLGYLAPEY--AMWGKVSESCDVYSFGILLLELVTGRK 904
>AV421705
Length = 341
Score = 143 bits (360), Expect = 5e-35
Identities = 65/114 (57%), Positives = 87/114 (76%)
Frame = +3
Query: 90 NGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICD 149
NG++FI+EVA+I R H+N+V L+GYC + KRAL+YEFMP GSLDK+I+ E
Sbjct: 9 NGQDFISEVATIGRIHHINVVRLVGYCVDGKKRALVYEFMPNGSLDKYIFSKE---GSAW 179
Query: 150 FDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLA 203
++ ++ I++GIA+G+ YLHQGC +ILH DIKP NILLD+DF PK+SDFGLA
Sbjct: 180 LSYDQIYEISLGIARGVAYLHQGCDMQILHFDIKPHNILLDKDFIPKVSDFGLA 341
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 142 bits (357), Expect = 1e-34
Identities = 72/167 (43%), Positives = 105/167 (62%), Gaps = 3/167 (1%)
Frame = +3
Query: 42 RRYSYAEVKMITNYFR--EKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEV 98
+ +SY + T F KLG+GG+G V+K L + R++AVK +S G +FINE
Sbjct: 201 KTFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLNDGREIAVKKLSRRSNQGRTQFINEA 380
Query: 99 ASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRI 158
++R H N+VSL GYC +++ L+YE++P+ SLDK +++S+ + DW F I
Sbjct: 381 KLLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLLFRSQKKEQL---DWKRRFDI 551
Query: 159 AIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKI 205
G+A+GL YLH+ I+H DIK NILLDE + PKI+DFGLA+I
Sbjct: 552 ISGVARGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADFGLARI 692
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 140 bits (354), Expect = 3e-34
Identities = 85/273 (31%), Positives = 141/273 (51%), Gaps = 3/273 (1%)
Frame = +3
Query: 60 LGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFINEVASISRTSHMNIVSLLGYCYE 118
LG GG+G VY + +VA+K + G EF E+ +S+ H ++VSL+GYC E
Sbjct: 15 LGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYCEE 194
Query: 119 ENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRIL 178
+ L+Y+ M G+L + +YK++ P W I IG A+GL YLH G I+
Sbjct: 195 NTEMILVYDHMAYGTLREHLYKTQKP----PLPWKQRLEICIGAARGLHYLHTGAKYTII 362
Query: 179 HLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVS 238
H D+K NILLDE + K+SDFGL+K D+ +G+ GY+ PE F R ++
Sbjct: 363 HRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQ--QLT 536
Query: 239 YRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDLEQG--NTLLNCSTISEEE 296
+SDVYS+G+++ E++ R + ++ +W +G + +L+ +
Sbjct: 537 DKSDVYSFGVVLFEILCARPALNP-SLAKEQVSLAEWASHCYNKGILDQILDPYLKGKIA 713
Query: 297 NDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQ 329
+ +K ++ C+ + +RP M V+ L+
Sbjct: 714 PECFKKFAETAMKCVSDQGIERPSMGDVLWNLE 812
>BI418434
Length = 478
Score = 139 bits (350), Expect = 8e-34
Identities = 71/160 (44%), Positives = 101/160 (62%)
Frame = +2
Query: 49 VKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASISRTSHMN 108
+++ T F EK+G GG+G V++ L +S VAVK + G +EF EV++I H+N
Sbjct: 2 LQLATRGFSEKVGHGGFGTVFQGELSDSSLVAVKRLERPGGGEKEFRAEVSTIGNIQHVN 181
Query: 109 IVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEY 168
+V L G+C E + R L+YE+M G+L ++ K + C W+ F++AIG AKG+ Y
Sbjct: 182 LVRLRGFCSENSHRLLVYEYMHNGALSAYLRK----DGPC-LSWDVRFQVAIGTAKGIAY 346
Query: 169 LHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQR 208
LH+ ILH DIKP+NILLD DF K+SDFGLAK+ R
Sbjct: 347 LHEELRCCILHCDIKPENILLDSDFTAKVSDFGLAKLIGR 466
>BP032158
Length = 555
Score = 138 bits (348), Expect = 1e-33
Identities = 78/183 (42%), Positives = 112/183 (60%), Gaps = 3/183 (1%)
Frame = +2
Query: 44 YSYAEVKMITNYF--REKLGQGGYGVVYKASLYNSRQVAVKVIS-ETKGNGEEFINEVAS 100
+S ++K TN F K+G+GG+G VYK L +AVK +S ++K EFINE+
Sbjct: 14 FSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSSKSKQGNREFINEIGM 193
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAI 160
IS H N+V L G C E N+ L+YE+M SL + ++ +E + +W T +I +
Sbjct: 194 ISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKL--NLNWRTRMKICV 367
Query: 161 GIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARG 220
GIAKGL YLH+ +I+H DIK N+LLD+D KISDFGLAK+ + +++ +S A G
Sbjct: 368 GIAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHISTRIA-G 544
Query: 221 TIG 223
TIG
Sbjct: 545 TIG 553
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 137 bits (346), Expect = 2e-33
Identities = 74/188 (39%), Positives = 111/188 (58%), Gaps = 3/188 (1%)
Frame = +1
Query: 43 RYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEVASI 101
+YSY +++ T F LGQG +G VYKA++ VAVKV++ GE EF EV +
Sbjct: 196 KYSYKDIQKATQNFTTILGQGSFGTVYKATMPTGEVVAVKVLAPNSKQGEHEFQTEVHLL 375
Query: 102 SRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIG 161
R H N+V+L+G+C ++ +R L+Y+FM GSL +Y E + W+ +IA+
Sbjct: 376 GRLHHRNLVNLVGFCVDKGQRILVYQFMSNGSLANLLYGEEK-----ELSWDERLQIAMD 540
Query: 162 IAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAK--ICQRKDSIVSILGAR 219
I+ G+EYLH+G ++H D+K NILLD+ ++DFGL+K I ++S G +
Sbjct: 541 ISHGIEYLHEGAVPPVIHRDLKSANILLDDSMRAMVADFGLSKEEIFDGRNS-----GLK 705
Query: 220 GTIGYMAP 227
GT GYM P
Sbjct: 706 GTYGYMDP 729
>BP085623
Length = 528
Score = 127 bits (318), Expect = 4e-30
Identities = 59/123 (47%), Positives = 83/123 (66%)
Frame = -3
Query: 225 MAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDLEQGN 284
MAPE+F + GGVSY+ DVYS+GML++EM RKN + +S++YFP WIY L + N
Sbjct: 526 MAPELFYKNIGGVSYKXDVYSFGMLLMEMASNRKNLNPHVEHSSQLYFPFWIYDQLGKQN 347
Query: 285 TLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLY 344
+ + EEE + +K+ +V+LWCIQ KP DRP M +V+EML+G + ++ PPKP LY
Sbjct: 346 D-IELEDVKEEEVKIAKKMIMVALWCIQLKPDDRPSMKEVVEMLEGDVENLEMPPKPSLY 170
Query: 345 SPE 347
E
Sbjct: 169 PHE 161
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 127 bits (318), Expect = 4e-30
Identities = 90/302 (29%), Positives = 153/302 (49%), Gaps = 9/302 (2%)
Frame = +1
Query: 44 YSYAEVKMITNYFR--EKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASI 101
+SY E+ TN F K+GQGG+G VY A L ++ A+K + EF+ E+ +
Sbjct: 1051 FSYQELAKATNNFSLDNKIGQGGFGAVYYAEL-RGKKTAIKKMDVQAST--EFLCELKVL 1221
Query: 102 SRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIG 161
+ H+N+V L+GYC E L+YE + G+L ++++ S W++ +IA+
Sbjct: 1222 THVHHLNLVRLIGYCVE-GSLFLVYEHIDNGNLGQYLHGSGKE----PLPWSSRVQIALD 1386
Query: 162 IAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGT 221
A+GLEY+H+ +H D+K NIL+D++ K++DFGL K+ + +S + GT
Sbjct: 1387 AARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQ-TRLVGT 1563
Query: 222 IGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSE-----MYFPDWI 276
GYM PE +G +S + DVY++G+++ E+I + G +E F + +
Sbjct: 1564 FGYMPPEY--AQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESKGLVALFEEAL 1737
Query: 277 YKD--LEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSS 334
K + L++ D + KI + C + P RP M ++ L LSS
Sbjct: 1738 NKSDPCDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVALM-TLSS 1914
Query: 335 VS 336
++
Sbjct: 1915 LT 1920
>BP064337
Length = 485
Score = 125 bits (313), Expect = 1e-29
Identities = 59/99 (59%), Positives = 75/99 (75%)
Frame = -1
Query: 273 PDWIYKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPL 332
PDWIYKDLEQG+ N I+EEE++M RK+ LVSLWCI T+ S+RP MNKV+EML+G L
Sbjct: 485 PDWIYKDLEQGDVHTNFLVITEEEHEMARKMILVSLWCIPTRSSERPSMNKVVEMLEGTL 306
Query: 333 SSVSYPPKPVLYSPERPELQVSDMSSSDLYETNSVTVSK 371
SV YPPKP+L+SPE+ L SD++S ET+S+T K
Sbjct: 305 ESVPYPPKPILHSPEKLLLPNSDLTSGHTLETDSMTTEK 189
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,278,373
Number of Sequences: 28460
Number of extensions: 90590
Number of successful extensions: 907
Number of sequences better than 10.0: 296
Number of HSP's better than 10.0 without gapping: 712
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 717
length of query: 371
length of database: 4,897,600
effective HSP length: 92
effective length of query: 279
effective length of database: 2,279,280
effective search space: 635919120
effective search space used: 635919120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC147875.1


