

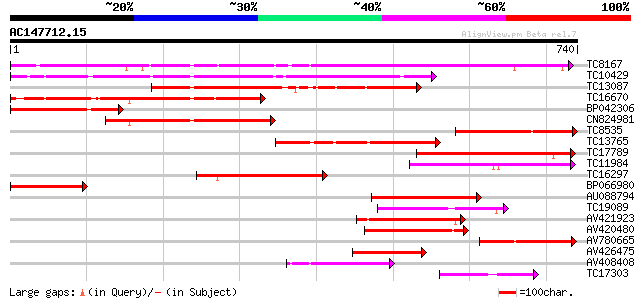
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147712.15 + phase: 0 /pseudo/partial
(740 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8167 similar to PIR|JC7519|JC7519 subtilisin-like serine prote... 478 e-135
TC10429 similar to UP|Q9ZR46 (Q9ZR46) P69C protein, partial (52%) 344 3e-95
TC13087 similar to UP|Q9FIF8 (Q9FIF8) Serine protease-like prote... 244 3e-65
TC16670 similar to PIR|JC7519|JC7519 subtilisin-like serine prot... 233 8e-62
BP042306 219 9e-58
CN824981 205 2e-53
TC8535 weakly similar to UP|Q9FJF3 (Q9FJF3) Serine protease-like... 166 1e-41
TC13765 similar to UP|Q9LLL8 (Q9LLL8) Subtilisin-type serine end... 165 3e-41
TC17789 similar to PIR|T05768|T05768 subtilisin-like proteinase ... 163 8e-41
TC11984 similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, ... 162 2e-40
TC16297 weakly similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type pro... 160 9e-40
BP066980 153 1e-37
AU088794 152 2e-37
TC19089 similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C... 137 6e-33
AV421923 127 8e-30
AV420480 126 1e-29
AV780665 116 1e-26
AV426475 108 4e-24
AV408408 105 2e-23
TC17303 weakly similar to UP|O82440 (O82440) Subtilisin-like pro... 94 6e-20
>TC8167 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (65%)
Length = 2760
Score = 478 bits (1231), Expect = e-135
Identities = 288/758 (37%), Positives = 431/758 (55%), Gaps = 22/758 (2%)
Frame = +3
Query: 1 SYIVYIGSHSHGP-NPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAV 59
+YIV++ H+ P+ D +A+ +L E + + + Y+Y+ NGFAA
Sbjct: 147 TYIVHMNHHTKPQIYPTRRDWYTASLRSLSLTTDS----ETSDDPLLYAYDTAYNGFAAS 314
Query: 60 LEVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTI 119
L+ ++A + +V+ ++E+ + L TTR+ +FLGL+ G+ E + I
Sbjct: 315 LDEQQAQTLLGSDSVLGLYEDTLYHLHTTRTPQFLGLDTQTGLWEGHRTLELDQASRDVI 494
Query: 120 IANIDSGVSPESKSFSDDGMGPVPSRWRGICQ----LDNFHCNRKLIGARFYSQGY---- 171
I +D+GV PES SF+D GM +PSRWRG C+ + CNRKLIGAR +S+G+
Sbjct: 495 IGVLDTGVWPESPSFNDAGMPEIPSRWRGECENATDFSSSLCNRKLIGARSFSRGFHMAA 674
Query: 172 --ESKFGRLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAY 229
+ FG+ + + RD GHGT T S A G+ V A++ G A+GTA+G +P++ VA Y
Sbjct: 675 GNDGGFGKEREPP-SPRDSDGHGTHTASTAAGSHVGNASLLGYASGTARGMAPQARVATY 851
Query: 230 KVCWLGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENG 289
KVCW C +DI+ + AI DGVD++S SLG S +F D I+IGAF A+E G
Sbjct: 852 KVCWSDG----CFASDILAGMDRAIRDGVDVLSLSLGGGSGP-YFRDTIAIGAFAAMERG 1016
Query: 290 VIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNE 349
+ V GNSGP ++ NVAPWL +V A T+DR+F + LG+K G SL +G
Sbjct: 1017IFVSCSAGNSGPSKASLANVAPWLMTVGAGTLDRDFPASALLGNKKRFAGVSLYSG--KG 1190
Query: 350 KFYSLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGG 409
V V +K N + +C GSLDP V+GK++ C R L+ V + G
Sbjct: 1191MGAEPVGLVYSKGSN---QSGILCLPGSLDPAVVRGKVVLCD-RGLNARVEKGKVVKEAG 1358
Query: 410 SIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVK 469
IG++L N G +++A +HLLP + G+ + Y+ + P A ++ + T + V+
Sbjct: 1359GIGMILTNTAANGEELVADSHLLPAVAVGRIVGDQIREYVTSDPNPTAVLSFSGTVLNVR 1538
Query: 470 PAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGT 529
P+PV+A+ SSRGPN I ILKPD+ PGV+IL + AI P+GL D++ +NI SGT
Sbjct: 1539PSPVVAAFSSRGPNMITKQILKPDVIGPGVNILAGWSEAIGPSGLPQDSRKSQFNIMSGT 1718
Query: 530 SISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQS-KEDATPFGYGA 588
S+SCPH+S + ALLK +P+WSP+A KSA+MTT + N + P++D + E +TP+ +GA
Sbjct: 1719SMSCPHISGLGALLKAAHPDWSPSAIKSALMTTAYVHDNTNSPLRDAAGGEFSTPWAHGA 1898
Query: 589 GHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYI--CPKSYNMLDFNYP 646
GH+ P+ A+ PGLVYD DY+ FLC+ Y+ +++ ++ + K + NYP
Sbjct: 1899GHVNPQKALSPGLVYDAKARDYIAFLCSLDYSPDHLQLIVKRAGVNCSRKLSDPGQLNYP 2078
Query: 647 SITVPNLGKHF----VQEVTRTVTNVGSPGT-YRVQVNEPHGIFVLIKPRSLTFNEVGEK 701
S +V F V TRTVTNVG G+ Y V V+ P + + + P L F +VGE+
Sbjct: 2079SFSVVFDSVVFDAKKVVRYTRTVTNVGEAGSVYDVVVDGPSMVGITVYPTKLEFGKVGER 2258
Query: 702 KTFKIIFKVTKPTSSGYV---FGHLLWSDGRHKVMSPL 736
K + + F K S V FG + W + +H+V SP+
Sbjct: 2259KRYTVTFVSKKGASDDLVRSAFGSITWKNEQHQVRSPV 2372
>TC10429 similar to UP|Q9ZR46 (Q9ZR46) P69C protein, partial (52%)
Length = 1742
Score = 344 bits (883), Expect = 3e-95
Identities = 210/555 (37%), Positives = 307/555 (54%)
Frame = +3
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
YI+++ + DL+S +++ L L S E+ I YSY + GFAA L
Sbjct: 153 YIIHVTGPEGKMLTESEDLESW---YHSFLPPTLKSSEEQPRVI-YSYKNVLRGFAASLT 320
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
EE + + + + + + ++ +FLGL+ + GV W++ +G+G II
Sbjct: 321 QEELSAVERKMALFQLILRGCYIVKPLIPPKFLGLQQDTGV------WKESNFGKGVIIG 482
Query: 122 NIDSGVSPESKSFSDDGMGPVPSRWRGICQLDNFHCNRKLIGARFYSQGYESKFGRLNQS 181
+DSG++P SFSD G+ P P +W+G C L+ CN KLIGAR ++ E+ G+ ++
Sbjct: 483 VLDSGITPGHPSFSDVGIPPPPPKWKGRCDLNVTACNNKLIGARAFNLAAEAMNGKKAEA 662
Query: 182 LYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IEC 241
D GHGT T S A G FV+ A V G A GTA G +P +H+A YKVC+ +C
Sbjct: 663 PI---DEDGHGTHTASTAAGAFVNYAEVLGNAKGTAAGMAPHAHLAIYKVCFGE----DC 821
Query: 242 TDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGP 301
++DI+ A + A+ DGVD+IS SLG + P FF D +IGAF A++ G+ V GNSGP
Sbjct: 822 PESDILAALDAAVEDGVDVISISLGLSEPPPFFNDSTAIGAFAAMQKGIFVSCAAGNSGP 1001
Query: 302 KFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAK 361
++ N APW+ +V ASTIDR V+ +LG+ G S+ P+ +L+ A
Sbjct: 1002FNSSIVNAAPWILTVGASTIDRRIVATAKLGNGQEFDGESVFQ--PSSFTPTLLPL--AY 1169
Query: 362 VGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDKQR 421
G E++ C GSLD + +GK++ C + EE G ++L ND+
Sbjct: 1170AGKNGKEESAFCANGSLDDSAFRGKVVLCERGGGIARIAKGEEVKRAGGAAMILMNDETN 1349
Query: 422 GNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRG 481
+ A H LP +H++Y G + +YI +T TP A + T +G AP +AS SSRG
Sbjct: 1350AFSLSADVHALPATHVSYAAGIEIKAYINSTATPTATILFKGTVIGNSLAPAVASFSSRG 1529
Query: 482 PNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVA 541
PN P ILKPDI PGV+IL A+ +S S + + +NI SGTS+SCPH+S I A
Sbjct: 1530PNLPSPGILKPDIIGPGVNILAAWPFPLS----NSTDSKLTFNIESGTSMSCPHLSGIAA 1697
Query: 542 LLKTIYPNWSPAAFK 556
LLK+ +P+WSPAA K
Sbjct: 1698LLKSSHPHWSPAAIK 1742
>TC13087 similar to UP|Q9FIF8 (Q9FIF8) Serine protease-like protein, partial
(31%)
Length = 1036
Score = 244 bits (624), Expect = 3e-65
Identities = 139/356 (39%), Positives = 215/356 (60%), Gaps = 4/356 (1%)
Frame = +2
Query: 186 RDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTDAD 245
RD+ GHG+ T S A GN V GA+ +GLA GTA+G P + +AAYKVC+ C DA+
Sbjct: 26 RDIEGHGSHTASTAAGNTVIGASFYGLAQGTARGAVPSARIAAYKVCYPNH---GCDDAN 196
Query: 246 IMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGT 305
I+ AF+DAI+DGV+I+S SLG + D ++IG+FHA+E G++ + GNSGP +
Sbjct: 197 ILAAFDDAIADGVNILSVSLGTNYCVDLLSDTVAIGSFHAMERGILTLQAAGNSGPASPS 376
Query: 306 VTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNA 365
+ +VAPWL +VAASTIDR F+ + LG+ ++G S++ + N + + NA
Sbjct: 377 ICSVAPWLLTVAASTIDRQFIDKVLLGNGMTLIGRSVNAVVSNGTKFPIAK------WNA 538
Query: 366 TIEDAK----ICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDKQR 421
+ E K +C+ L VKGK++ C +G ++ +E A G++G + +
Sbjct: 539 SDERCKGFSDLCQ--CLGSESVKGKVVLC-----EGRMH-DEAAYLDGAVGSITEDLTNP 694
Query: 422 GNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRG 481
ND+ ++ P+ ++N D V +Y + K P A + K++ + AP + S SSRG
Sbjct: 695 DNDV-SFVTYFPSVNLNPKDYAIVQNYTNSAKNPKAEILKSEI-FKDRSAPKVVSFSSRG 868
Query: 482 PNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVS 537
PNP+ P I+KPD++APGVDIL A+ +P+ +SD + + YN+ SGTS++CPHV+
Sbjct: 869 PNPLIPEIMKPDVSAPGVDILAAWSPEAAPSEYSSDKRKVRYNVVSGTSMACPHVA 1036
>TC16670 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (42%)
Length = 1183
Score = 233 bits (594), Expect = 8e-62
Identities = 136/339 (40%), Positives = 206/339 (60%), Gaps = 6/339 (1%)
Frame = +2
Query: 1 SYIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVL 60
+YIV++ S++ + + H S L S + E + Y+Y+ I+GF+ L
Sbjct: 230 TYIVHVAK---------SEMPESFEHHAAWYESSLQSVSDSAEML-YTYDNAIHGFSTRL 379
Query: 61 EVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTII 120
EEA + V++V + EL TTR+ FLGL+ + + P G GE I+
Sbjct: 380 TPEEARLLEAQTGVLAVLPERKFELHTTRTPLFLGLDKSADMFPAS-----GAGGE-VIV 541
Query: 121 ANIDSGVSPESKSFSDDGMGPVPSRWRGICQLD-NF---HCNRKLIGARFYSQGYESKFG 176
+D+GV PESKSF D G GPVPS W+G C+ NF +CN+KLIGAR++S+G E+ G
Sbjct: 542 GVLDTGVWPESKSFDDTGFGPVPSTWKGECESGTNFTAANCNKKLIGARYFSKGAEAMLG 721
Query: 177 RLNQSLYNA--RDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWL 234
++++ + RD GHGT T S A G+ VSGA++FG A GTA+G +P + VAAYKVCW
Sbjct: 722 PIDETAESKSPRDDDGHGTHTASTAAGSVVSGASLFGYAAGTARGMAPHARVAAYKVCWK 901
Query: 235 GTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVA 294
G C +DI+ A + AI+D V+++S SLG +++ D ++IGAF A+E G++V
Sbjct: 902 GG----CFSSDILAAIDKAIADNVNVLSLSLGG-GMADYYRDSVAIGAFAAMEKGILVSC 1066
Query: 295 GGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGD 333
GN+GP +++NVAPW+ +V A T+DR+F + ++LG+
Sbjct: 1067SAGNAGPSAYSLSNVAPWITTVGAGTLDRDFPALVELGN 1183
>BP042306
Length = 549
Score = 219 bits (559), Expect = 9e-58
Identities = 106/147 (72%), Positives = 125/147 (84%)
Frame = +3
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
YIVY+G+HSHGP PS+ DL++AT SHY+LLGS LGSHEKAKEAI YSYNKHINGFAA+LE
Sbjct: 111 YIVYLGAHSHGPTPSSVDLETATSSHYDLLGSVLGSHEKAKEAILYSYNKHINGFAALLE 290
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
EEAA+IAK+P VVSVF +K H+L TTRSWEFLGL N G +S W+KGR+GE TIIA
Sbjct: 291 EEEAARIAKNPKVVSVFVSKEHKLHTTRSWEFLGLRGNGG----NSAWQKGRFGENTIIA 458
Query: 122 NIDSGVSPESKSFSDDGMGPVPSRWRG 148
NID+GV PES+SFSD G+G VP++WRG
Sbjct: 459 NIDTGVWPESQSFSDKGIGSVPAKWRG 539
>CN824981
Length = 726
Score = 205 bits (521), Expect = 2e-53
Identities = 110/229 (48%), Positives = 151/229 (65%), Gaps = 6/229 (2%)
Frame = +1
Query: 125 SGVSPESKSFSDDGMGPVPSRWRGICQLDN----FHCNRKLIGARFYSQGYESKFGRLNQ 180
+GV PE KS D G+ PVPS W+G C+ N CNRKLIGARF+S+GYE+ G ++
Sbjct: 1 AGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRKLIGARFFSKGYEATLGPIDV 180
Query: 181 SLYN--ARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI* 238
S + ARD GHG+ TL+ A G+ V+GA++FGLA+GTA+G + ++ VAAYKVCWLG
Sbjct: 181 STESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLG--- 351
Query: 239 IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGN 298
C +DI + AI DGV+IIS S+G +S ++F D I+IGAF A +G++V GN
Sbjct: 352 -GCFSSDIAAGIDKAIEDGVNIISMSIGGSS-ADYFRDIIAIGAFTANSHGILVSTSAGN 525
Query: 299 SGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLP 347
GP +++N APW+ +V A TIDR+F +Y+ LG+ G SL G P
Sbjct: 526 GGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKP 672
>TC8535 weakly similar to UP|Q9FJF3 (Q9FJF3) Serine protease-like protein,
partial (10%)
Length = 780
Score = 166 bits (420), Expect = 1e-41
Identities = 83/161 (51%), Positives = 111/161 (68%), Gaps = 2/161 (1%)
Frame = +2
Query: 582 TPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFS-RKPYICPKSYNM 640
TPF YG+GH+QP A+DPGLVYDL+IVDYLNFLCA GY+Q + + K + C S+++
Sbjct: 2 TPFAYGSGHVQPNSAIDPGLVYDLSIVDYLNFLCASGYDQQLISALNFNKTFTCSGSHSI 181
Query: 641 LDFNYPSITVPNLGKHFVQEVTRTVTNVGSPGTYRVQVNEPHGIFVLIKPRSLTFNEVGE 700
D NYPSIT+PN+ + V VTRTVTNVG P Y P G +++ P SL+F ++GE
Sbjct: 182 TDLNYPSITLPNIRSNVV-NVTRTVTNVGPPSKYFANAQLP-GHKIVVVPNSLSFKKIGE 355
Query: 701 KKTFKIIFKVTKPTS-SGYVFGHLLWSDGRHKVMSPLVVKH 740
KKTF++I + T T Y+FG L W++GRH V SP+ V+H
Sbjct: 356 KKTFQVIVQATSVTQRRKYLFGKLQWTNGRHIVRSPITVRH 478
>TC13765 similar to UP|Q9LLL8 (Q9LLL8) Subtilisin-type serine endopeptidase
XSP1, partial (20%)
Length = 635
Score = 165 bits (417), Expect = 3e-41
Identities = 91/216 (42%), Positives = 133/216 (61%)
Frame = +1
Query: 347 PNEKFYSLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAI 406
P K YSL++ +DA + + +DA C SL PNKVKGK+++C L G E
Sbjct: 1 PKRKEYSLINGIDAAKDSKSKDDAGFCYEDSLQPNKVKGKLVYCKL----GNWGTEGVVK 168
Query: 407 SGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEV 466
G IG ++ +D+ +A + P + +N+T GE V +YIK+T++P A + K E
Sbjct: 169 KFGGIGSIMESDQYPD---LAQIFMAPATILNHTIGESVTNYIKSTRSPSAVIYKTHEEK 339
Query: 467 GVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIG 526
PAP +A+ SSRGPNP +LKPDI APG+DIL +Y S TG D Q+ +++
Sbjct: 340 C--PAPFVATFSSRGPNPGSHNVLKPDIAAPGIDILASYTLRKSITGSEGDTQFSEFSLL 513
Query: 527 SGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTT 562
SGTS++CPHV+ + A +K+ +PNW+PAA +SAI+TT
Sbjct: 514 SGTSMACPHVAGVAAYVKSFHPNWTPAAIRSAIITT 621
>TC17789 similar to PIR|T05768|T05768 subtilisin-like proteinase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (14%)
Length = 940
Score = 163 bits (413), Expect = 8e-41
Identities = 91/219 (41%), Positives = 136/219 (61%), Gaps = 11/219 (5%)
Frame = +2
Query: 531 ISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSK-EDATPFGYGAG 589
++CPH+S ALLK+ +P+WSPAA +SA+MTT T+ N + + D++ +TP+ +GAG
Sbjct: 2 MACPHISGAAALLKSAHPDWSPAAVRSAMMTTATVLDNRKQILTDEATGNSSTPYDFGAG 181
Query: 590 HIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICP-KSYNMLDFNYPS- 647
H+ LAMDPGLVYD+ DY+NFLC GY +++ +R P CP + + + NYPS
Sbjct: 182 HVNLGLAMDPGLVYDITGADYVNFLCGIGYGPRVIQVITRAPVSCPARKPSPENLNYPSF 361
Query: 648 ITVPNLGKHFVQEVT--RTVTNVG-SPGTYRVQV-NEPHGIFVLIKPRSLTFNEVGEKKT 703
+ + +G + T RTVTNVG + YRV V ++ G+ V +KP L F+E +K++
Sbjct: 362 VAMFPVGSKGLTTKTFIRTVTNVGPANSVYRVSVESQMKGVTVTVKPSRLVFSEAVKKRS 541
Query: 704 FKIIF----KVTKPTSSGYVFGHLLWSDGRHKVMSPLVV 738
+ + + K SG VFG L W+DG+H V SP+VV
Sbjct: 542 YVVTVAAETRFLKMGPSGAVFGSLTWTDGKHVVRSPIVV 658
>TC11984 similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, partial
(30%)
Length = 966
Score = 162 bits (409), Expect = 2e-40
Identities = 90/225 (40%), Positives = 129/225 (57%), Gaps = 9/225 (4%)
Frame = +1
Query: 523 YNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDAT 582
YNI SGTS+SCPHVS + +K+ + WS +A KSAIMT+ T N P+ S AT
Sbjct: 58 YNIISGTSMSCPHVSGLAGSIKSRHTTWSASAIKSAIMTSATQNNNLKAPMTTDSGSVAT 237
Query: 583 PFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSR---KPYICPK--- 636
P+ YGAG + + PGLVY+ + DYLN+LC G N T +K+ SR + CP+
Sbjct: 238 PYDYGAGEMTTSASFQPGLVYETSTTDYLNYLCYIGLNVTTIKVISRTVPDSFTCPQDSS 417
Query: 637 SYNMLDFNYPSITVPNLGKHFVQEVTRTVTNVGSPG--TYRVQVNEPHGIFVLIKPRSLT 694
+ ++ + NYPSI + N V+RTVTNVG Y V+ P G+ + + P L
Sbjct: 418 ADHVSNINYPSIAITNFNGKGAINVSRTVTNVGEEDETVYSPIVDAPSGVNIKLIPEKLP 597
Query: 695 FNEVGEKKTFKIIFKVTKPTS-SGYVFGHLLWSDGRHKVMSPLVV 738
F + +K ++ +IF +T TS +FG L WS+G++ V SP V+
Sbjct: 598 FTKSSQKLSYPVIFSLTSSTSLKEDLFGSLAWSNGKYIVRSPFVL 732
>TC16297 weakly similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease,
partial (10%)
Length = 574
Score = 160 bits (404), Expect = 9e-40
Identities = 84/174 (48%), Positives = 112/174 (64%), Gaps = 3/174 (1%)
Frame = +2
Query: 245 DIMQAFEDAISDGVDIISCSLGQTS---PKEFFEDGISIGAFHAIENGVIVVAGGGNSGP 301
D++ A + AI+DGVD+ S S G S +E F D +SIG+ HA+ ++VVA GN GP
Sbjct: 2 DVLAAIDQAITDGVDVNSVSAGGRSIISAEEIFTDEVSIGSVHALAKNILVVASAGNDGP 181
Query: 302 KFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAK 361
GTV NVAPW+F+VAASTIDR F S + LG+ I G SL LP + +SL+ S DAK
Sbjct: 182 ALGTVLNVAPWVFTVAASTIDREFSSTITLGNNQQITGASLFVNLPPNQLFSLILSTDAK 361
Query: 362 VGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVL 415
+ NAT DA++C+ G+LDP KVKGK++ C V EA+S G+ G+VL
Sbjct: 362 LANATNRDAQLCRPGTLDPAKVKGKVVVCTRGGKIKSVAEGSEALSSGAKGMVL 523
>BP066980
Length = 472
Score = 153 bits (386), Expect = 1e-37
Identities = 73/100 (73%), Positives = 84/100 (84%)
Frame = +3
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
YIVY+G+HSHGP PS+ DL++AT SHY+LLGS LGSHEKAKEAI YSYNKHINGFAA+LE
Sbjct: 168 YIVYLGAHSHGPTPSSVDLETATSSHYDLLGSVLGSHEKAKEAILYSYNKHINGFAALLE 347
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYG 101
EEAA+I K+P VVSVF +K H + T RSWEFLGL N G
Sbjct: 348 EEEAARIGKNPKVVSVFVSKEH*IDTNRSWEFLGLRGNGG 467
>AU088794
Length = 433
Score = 152 bits (383), Expect = 2e-37
Identities = 74/144 (51%), Positives = 99/144 (68%), Gaps = 1/144 (0%)
Frame = +2
Query: 473 VIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSIS 532
V+AS S+RGPNP P ILKPD+ PG++IL A+ P+G+ SD + +NI SGTS++
Sbjct: 2 VVASFSARGPNPESPEILKPDVIXPGLNILAAWPDXXGPSGVPSDVRRTEFNILSGTSMA 181
Query: 533 CPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDAT-PFGYGAGHI 591
CPHVS + ALLK +P WSPAA KSA+MTT N + D+S + + F YG+GH+
Sbjct: 182 CPHVSGLAALLKAAHPXWSPAAIKSALMTTAYTVDNKGDAMLDESNGNVSLVFDYGSGHV 361
Query: 592 QPELAMDPGLVYDLNIVDYLNFLC 615
P AMDPGLVYD++ DY++FLC
Sbjct: 362 HPXKAMDPGLVYDISTYDYVDFLC 433
>TC19089 similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, partial
(16%)
Length = 600
Score = 137 bits (345), Expect = 6e-33
Identities = 74/176 (42%), Positives = 99/176 (56%), Gaps = 6/176 (3%)
Frame = +1
Query: 481 GPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIV 540
GPNPI P ILKPDI APGV +L A+ + + D + +PYN SGTS++CPH +A
Sbjct: 16 GPNPITPNILKPDIAAPGVTVLAAWSPLNPISEVEGDKRKLPYNAISGTSMACPHAAAAA 195
Query: 541 ALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELAMDPG 600
A +K+ +PNWSPA KSA+MTT T + P F YGAG I P A++PG
Sbjct: 196 AYVKSFHPNWSPAMIKSALMTTATPMNSALNP--------EAEFAYGAGQINPVKAVNPG 351
Query: 601 LVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYIC------PKSYNMLDFNYPSITV 650
LVYD+ DY+ FLC GY T ++ ++ C +YN+ + P I V
Sbjct: 352 LVYDITEQDYIQFLCGXGYTDTLLQNLTQHKRXCFGXSQXKXAYNLNLHHLPFIMV 519
>AV421923
Length = 463
Score = 127 bits (318), Expect = 8e-30
Identities = 67/154 (43%), Positives = 95/154 (61%), Gaps = 12/154 (7%)
Frame = +2
Query: 453 KTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPT 512
K P A ++ T G PAPV+A+ SSRGP+ + ++KPD+TAPGV IL A+ SP+
Sbjct: 2 KKPTASISFIGTRFG-DPAPVVAAFSSRGPSIVGLDVIKPDVTAPGVHILAAWPSKTSPS 178
Query: 513 GLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRP 572
L +D + + +NI SGTS+SCPHVS + AL+++++ +WSPAA KSA+MTT N P
Sbjct: 179 MLKNDKRRVLFNIISGTSMSCPHVSGVAALIRSVHRDWSPAAVKSALMTTAYTLNNKGAP 358
Query: 573 IKDQSKED------------ATPFGYGAGHIQPE 594
I D + A PF +G+GH+ PE
Sbjct: 359 ISDIGASNSISNSNSNHSGFAVPFAFGSGHVNPE 460
>AV420480
Length = 412
Score = 126 bits (317), Expect = 1e-29
Identities = 63/135 (46%), Positives = 90/135 (66%)
Frame = +3
Query: 464 TEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPY 523
T +G AP +A+ S+RGP+ P ILKPD+ APGV+I+ A+ + PT L D + + +
Sbjct: 9 TVIGNSRAPAVATFSARGPSFTNPSILKPDVVAPGVNIIAAWPQNLGPTSLPQDLRRVNF 188
Query: 524 NIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATP 583
++ SGTS+SCPHVS I AL+ + +P WSPAA KSAIMTT + + RPI D+ K A
Sbjct: 189 SVMSGTSMSCPHVSGIAALVHSAHPKWSPAAIKSAIMTTADVTDHMKRPILDEDK-PAGV 365
Query: 584 FGYGAGHIQPELAMD 598
F GAG++ P+ A++
Sbjct: 366 FAIGAGNVNPQRALN 410
>AV780665
Length = 505
Score = 116 bits (291), Expect = 1e-26
Identities = 61/127 (48%), Positives = 84/127 (66%), Gaps = 1/127 (0%)
Frame = -2
Query: 614 LCAHGYNQTQMKMFSRKPYICPKSYNMLDFNYPSITVPNLGKHFVQEVTRTVTNVGSPGT 673
LCA GYN TQ+ + S PY C K++++L+ N PSITVP+L VTRT+ NVGS T
Sbjct: 501 LCALGYNDTQISVPSEGPYPCHKNFSLLNLNDPSITVPDLKGTVT--VTRTLKNVGSRAT 328
Query: 674 YRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTK-PTSSGYVFGHLLWSDGRHKV 732
Y V P+G+ + + P L FN VGE+K++K+ KV + T++ YVFG L+WSDG +
Sbjct: 327 YIAHVQHPNGVTISV*PTLLKFNHVGEEKSYKVKLKVKQGKTTNAYVFGKLIWSDGEPYL 148
Query: 733 MSPLVVK 739
SP+VVK
Sbjct: 147 RSPIVVK 127
>AV426475
Length = 300
Score = 108 bits (269), Expect = 4e-24
Identities = 51/97 (52%), Positives = 70/97 (71%)
Frame = +2
Query: 448 YIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIG 507
Y+ +++ P A + T + VKP+PV+A+ SSRGPN + P ILKPD+ APGV+IL + G
Sbjct: 8 YVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTG 187
Query: 508 AISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLK 544
AI PTGL D + + +NI SGTS+SCPHVS + A+LK
Sbjct: 188 AIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILK 298
>AV408408
Length = 429
Score = 105 bits (263), Expect = 2e-23
Identities = 56/141 (39%), Positives = 84/141 (58%)
Frame = +3
Query: 362 VGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDKQR 421
+ N T + +C +G+L P+KV GKI+ C R ++ V + G +G+VL N
Sbjct: 3 ISNTT--NGNLCMMGTLSPDKVAGKIVMCD-RGMNARVQKGQVVKGAGGLGMVLSNTAAN 173
Query: 422 GNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRG 481
G +++A AHLLPTS + G + Y+ + + T+VG++P+PV+A+ SSRG
Sbjct: 174 GEELVADAHLLPTSAVGEKAGNAIKKYLFSDPKATVKILFEGTKVGIQPSPVVAAFSSRG 353
Query: 482 PNPIQPIILKPDITAPGVDIL 502
PN I P ILKPD+ APGV+IL
Sbjct: 354 PNSITPQILKPDLIAPGVNIL 416
>TC17303 weakly similar to UP|O82440 (O82440) Subtilisin-like protease
(Fragment), partial (4%)
Length = 536
Score = 94.4 bits (233), Expect = 6e-20
Identities = 54/133 (40%), Positives = 78/133 (58%), Gaps = 3/133 (2%)
Frame = -3
Query: 561 TTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHG-Y 619
TT +GN + + +DATPF YGAG IQP LA+DP L+Y+L+I D+++ G Y
Sbjct: 510 TTKDYRGNPMQESDFKKLKDATPFAYGAGVIQPNLAVDPDLIYELDIKDHMDHFWHRGIY 331
Query: 620 NQTQMKMFSRKPYICPKSYNMLD--FNYPSITVPNLGKHFVQEVTRTVTNVGSPGTYRVQ 677
++ + K + N+L+ +NYPSI++PNL + Q TR +TNVGSPG Y V
Sbjct: 330 DKDESK----------STLNILNYKYNYPSISIPNLQFNHPQTATRVLTNVGSPGKYEVT 181
Query: 678 VNEPHGIFVLIKP 690
V P + V + P
Sbjct: 180 VEAPPQVRVEVDP 142
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,945,083
Number of Sequences: 28460
Number of extensions: 181028
Number of successful extensions: 963
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 900
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 907
length of query: 740
length of database: 4,897,600
effective HSP length: 97
effective length of query: 643
effective length of database: 2,136,980
effective search space: 1374078140
effective search space used: 1374078140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147712.15


