

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147712.11 + phase: 0
(476 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
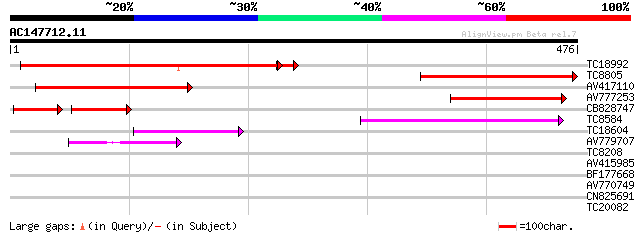
Score E
Sequences producing significant alignments: (bits) Value
TC18992 similar to UP|Q9LFZ2 (Q9LFZ2) F20N2.19, partial (18%) 370 e-107
TC8805 similar to GB|AAD28199.1|4704662|AF116860 uracil phosphor... 246 8e-66
AV417110 243 4e-65
AV777253 166 8e-42
CB828747 99 9e-38
TC8584 similar to UP|UPP_TOBAC (P93394) Uracil phosphoribosyltra... 72 2e-13
TC18604 homologue to UP|KPPR_MESCR (P27774) Phosphoribulokinase,... 57 7e-09
AV779707 48 3e-06
TC8208 similar to GB|AAL16268.1|16226812|AF428338 AT5g66680/MSN2... 29 1.9
AV415985 28 2.5
BF177668 27 7.3
AV770749 27 7.3
CN825691 27 9.5
TC20082 weakly similar to UP|AAR27072 (AAR27072) EIN3-binding F-... 27 9.5
>TC18992 similar to UP|Q9LFZ2 (Q9LFZ2) F20N2.19, partial (18%)
Length = 754
Score = 370 bits (950), Expect(2) = e-107
Identities = 183/223 (82%), Positives = 202/223 (90%), Gaps = 3/223 (1%)
Frame = +1
Query: 10 LMGSSSEVHFSGFHMD-GFEKREASIEQPTTSATDVYNQPFVIGVAGGAASGKKTVCDMI 68
LMGS+SEVHFSGFH+D G E+R+ EQPTTSATD+Y QPFVIGVAGGAASGK VCDMI
Sbjct: 46 LMGSASEVHFSGFHIDDGLERRKGGAEQPTTSATDMYKQPFVIGVAGGAASGKTAVCDMI 225
Query: 69 VQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDTEELLRVMDKLKHGEAVDI 128
+QQLHDQRVVLVNQDSFY++LTEEE RVQDYNFDHP+AFDT++LL V+DKLK G+AVDI
Sbjct: 226 IQQLHDQRVVLVNQDSFYHSLTEEELTRVQDYNFDHPEAFDTQQLLCVLDKLKRGQAVDI 405
Query: 129 PKYDFKSYKSDA--LRRVNPSDVIILEGILVFHDPRVRELMNMKIFVDTDADVRLARRIR 186
PKYDFK YK+D RR+NP+DVIILEGILVFHDPRVR LMNMKIFVDTDADVRLARRI+
Sbjct: 406 PKYDFKGYKNDVAPARRLNPADVIILEGILVFHDPRVRALMNMKIFVDTDADVRLARRIK 585
Query: 187 RDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIIIPRG 229
RDT+ RDIGA+LDQYSKFVKPAFDDFILPTKKYADIIIPRG
Sbjct: 586 RDTAVNSRDIGAVLDQYSKFVKPAFDDFILPTKKYADIIIPRG 714
Score = 35.0 bits (79), Expect(2) = e-107
Identities = 14/18 (77%), Positives = 16/18 (88%)
Frame = +2
Query: 225 IIPRGGDNHVAVDLIVQH 242
+ P GGDNHVA+DLIVQH
Sbjct: 701 LYPVGGDNHVAIDLIVQH 754
>TC8805 similar to GB|AAD28199.1|4704662|AF116860 uracil
phosphoribosyltransferase 1 {Arabidopsis thaliana;} ,
partial (71%)
Length = 685
Score = 246 bits (627), Expect = 8e-66
Identities = 122/131 (93%), Positives = 127/131 (96%)
Frame = +3
Query: 346 ACCKGIKIGKILIHREGDNGQQLIYEKLPNDISERHVLLLDPILGTGNSAVQAISLLIQK 405
ACCKGIKIGKILIHREGDNGQQLIYEKLP DIS+RHVLLLDPILGTGNSAVQAISLLI+K
Sbjct: 3 ACCKGIKIGKILIHREGDNGQQLIYEKLPMDISDRHVLLLDPILGTGNSAVQAISLLIRK 182
Query: 406 GVPESNIIFLNLISAPKGVHVVCKCFPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGT 465
GVPESNIIFLNLISAP GVHVVCK FP+IKIVTSEIEIGLN+DFRVIPGMGEFGDRYFGT
Sbjct: 183 GVPESNIIFLNLISAPLGVHVVCKKFPKIKIVTSEIEIGLNKDFRVIPGMGEFGDRYFGT 362
Query: 466 DDDEQVVVASQ 476
DDDEQ+VV SQ
Sbjct: 363 DDDEQLVVPSQ 395
>AV417110
Length = 398
Score = 243 bits (621), Expect = 4e-65
Identities = 118/132 (89%), Positives = 124/132 (93%)
Frame = +1
Query: 22 FHMDGFEKREASIEQPTTSATDVYNQPFVIGVAGGAASGKKTVCDMIVQQLHDQRVVLVN 81
FHMDGFEKREA+ EQPTTSATD+Y QPF+IGVAGGAASGKKTVCDMI+QQLHDQRVVLVN
Sbjct: 1 FHMDGFEKREANTEQPTTSATDMYKQPFIIGVAGGAASGKKTVCDMIIQQLHDQRVVLVN 180
Query: 82 QDSFYNNLTEEERARVQDYNFDHPDAFDTEELLRVMDKLKHGEAVDIPKYDFKSYKSDAL 141
QDSFYNNLTEEE RVQDYNFDHPDAFDTE+LL VMDKLK GEAVDIPKYDFKS K+D L
Sbjct: 181 QDSFYNNLTEEEVTRVQDYNFDHPDAFDTEQLLLVMDKLKRGEAVDIPKYDFKSCKTDVL 360
Query: 142 RRVNPSDVIILE 153
RRVNPSDVIILE
Sbjct: 361 RRVNPSDVIILE 396
>AV777253
Length = 592
Score = 166 bits (420), Expect = 8e-42
Identities = 79/97 (81%), Positives = 89/97 (91%)
Frame = -1
Query: 371 EKLPNDISERHVLLLDPILGTGNSAVQAISLLIQKGVPESNIIFLNLISAPKGVHVVCKC 430
EKLP DISERHVLLLDP+L TGNSA QAI LLIQKGVPES+IIFLNLISAP+G+H VCK
Sbjct: 592 EKLPKDISERHVLLLDPVLATGNSANQAIELLIQKGVPESHIIFLNLISAPEGIHCVCKR 413
Query: 431 FPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGTDD 467
FP +KIVTSEI++ LNE++RVIPG+GEFGDRYFGTDD
Sbjct: 412 FPSLKIVTSEIDVALNEEYRVIPGLGEFGDRYFGTDD 302
>CB828747
Length = 345
Score = 98.6 bits (244), Expect(2) = 9e-38
Identities = 47/50 (94%), Positives = 48/50 (96%)
Frame = +3
Query: 53 VAGGAASGKKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNF 102
VAGGAASGKKTVCDMI+QQLHDQRVVLVNQDSFYNNLTEEE RVQDYNF
Sbjct: 195 VAGGAASGKKTVCDMIIQQLHDQRVVLVNQDSFYNNLTEEEVTRVQDYNF 344
Score = 75.5 bits (184), Expect(2) = 9e-38
Identities = 36/41 (87%), Positives = 39/41 (94%)
Frame = +2
Query: 4 AKSAVDLMGSSSEVHFSGFHMDGFEKREASIEQPTTSATDV 44
AKSA+DLM SSSEVHFSGFHMDGFEKREA+ EQPTTSATD+
Sbjct: 71 AKSALDLMESSSEVHFSGFHMDGFEKREANTEQPTTSATDM 193
>TC8584 similar to UP|UPP_TOBAC (P93394) Uracil phosphoribosyltransferase
(UMP pyrophosphorylase) (UPRTase) , partial (95%)
Length = 1277
Score = 72.4 bits (176), Expect = 2e-13
Identities = 47/174 (27%), Positives = 90/174 (51%), Gaps = 3/174 (1%)
Frame = +2
Query: 295 LVVEHGLGHLPFTEKQVITPTGSVYTG-VDFCKRLCGVSVIRSGESMENALRACCKGIKI 353
L+ E LP ++ +P G +D + + + ++R+G ++ + K
Sbjct: 419 LMYEASRDWLPTISGEIQSPMGVASVEFIDPREPVAVIPILRAGLALAEHASSILPATKT 598
Query: 354 GKILIHREGDNGQQLIY-EKLPNDISE-RHVLLLDPILGTGNSAVQAISLLIQKGVPESN 411
+ + R+ + Q IY KLP+ +E V ++DP+L TG + V A++LL +GV
Sbjct: 599 YHLGLSRDEETLQPTIYLNKLPDKFAEGSKVFVVDPMLATGGTIVAALNLLKDRGVGNKQ 778
Query: 412 IIFLNLISAPKGVHVVCKCFPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGT 465
I ++ ++AP + + + FP + + T I+ +N+ +IPG+G+ GDR +GT
Sbjct: 779 IRVISAVAAPPALQKLSEQFPGLHVYTGIIDPTVNDKGFIIPGLGDAGDRSYGT 940
>TC18604 homologue to UP|KPPR_MESCR (P27774) Phosphoribulokinase,
chloroplast precursor (Phosphopentokinase) (PRKASE)
(PRK) , partial (45%)
Length = 724
Score = 57.0 bits (136), Expect = 7e-09
Identities = 27/92 (29%), Positives = 52/92 (56%)
Frame = +2
Query: 105 PDAFDTEELLRVMDKLKHGEAVDIPKYDFKSYKSDALRRVNPSDVIILEGILVFHDPRVR 164
P A D + + + +K G +V P Y+ + D + P ++++EG+ +DPRVR
Sbjct: 446 PKANDFDLMYEQVKAIKEGISVQKPIYNHVTGLLDPPELIKPPKILVIEGLHPMYDPRVR 625
Query: 165 ELMNMKIFVDTDADVRLARRIRRDTSEKDRDI 196
EL++ I++D +V+ A +I+RD +E+ +
Sbjct: 626 ELLDFSIYLDISNEVKFAWKIQRDMAERGHSL 721
>AV779707
Length = 552
Score = 48.1 bits (113), Expect = 3e-06
Identities = 31/95 (32%), Positives = 49/95 (50%)
Frame = +1
Query: 50 VIGVAGGAASGKKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFD 109
++GVAG + +GK + I+ + V+ ++ YN + +R+ D NFD P D
Sbjct: 280 LVGVAGPSGAGKTVFTEKILNFMPSIAVITMDN---YN-----DASRIIDGNFDDPRWTD 435
Query: 110 TEELLRVMDKLKHGEAVDIPKYDFKSYKSDALRRV 144
+ LL + LK G++V +P YDFKS R V
Sbjct: 436 YDTLLENIQGLKAGKSVQVPIYDFKSSSRIGYRTV 540
>TC8208 similar to GB|AAL16268.1|16226812|AF428338 AT5g66680/MSN2_7
{Arabidopsis thaliana;} , partial (67%)
Length = 928
Score = 28.9 bits (63), Expect = 1.9
Identities = 24/85 (28%), Positives = 39/85 (45%), Gaps = 7/85 (8%)
Frame = +2
Query: 226 IPRGGDNHVAVDL----IVQHIRTKLGQHDLCKIYPNLYVIHSTFQI---RGMHTLIRDA 278
+ G D VA D +++ I T+ G D + + V HS + + G HTLI
Sbjct: 317 VDSGHDLIVAADSNASDLIREIATECGV-DFDEDLSAMVVDHSGYAVSSTEGDHTLIASD 493
Query: 279 QITKHDFVFYSDRLIRLVVEHGLGH 303
+ K D + S ++ V+ G+GH
Sbjct: 494 DLIKSDVILGSKKIEAPVLFQGIGH 568
>AV415985
Length = 277
Score = 28.5 bits (62), Expect = 2.5
Identities = 21/64 (32%), Positives = 26/64 (39%), Gaps = 2/64 (3%)
Frame = +1
Query: 53 VAGGAASGKKTVCDMIVQQLHDQR--VVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDT 110
V G A SGK T C + Q R + +VN D NFD+P A D
Sbjct: 115 VIGPAGSGKSTYCSSLHQHCETTRRTIHVVNLDPAAE-------------NFDYPVAMDV 255
Query: 111 EELL 114
EL+
Sbjct: 256 RELI 267
>BF177668
Length = 301
Score = 26.9 bits (58), Expect = 7.3
Identities = 14/34 (41%), Positives = 21/34 (61%)
Frame = -1
Query: 386 DPILGTGNSAVQAISLLIQKGVPESNIIFLNLIS 419
D + +G++AV SL + G+PE + I NLIS
Sbjct: 298 DSPIASGDAAVCGDSLFREVGIPEISFIKCNLIS 197
>AV770749
Length = 348
Score = 26.9 bits (58), Expect = 7.3
Identities = 12/31 (38%), Positives = 16/31 (50%)
Frame = -2
Query: 197 GAILDQYSKFVKPAFDDFILPTKKYADIIIP 227
G ++ K V PA DDFI P +I+P
Sbjct: 110 GVVITLLCKIVAPALDDFIYPIPVIYILILP 18
>CN825691
Length = 590
Score = 26.6 bits (57), Expect = 9.5
Identities = 24/90 (26%), Positives = 40/90 (43%), Gaps = 10/90 (11%)
Frame = +2
Query: 349 KGIKIGKILIHREGDNGQQLIYEKLPNDI-----SERHVLLLDPILGTGNSAVQAISLLI 403
KGI + +++ G+ Q++YE+ DI E VLL D + V+
Sbjct: 206 KGINVRVLVVINPGNPTGQVLYEENQRDIVKFCKQEGLVLLADEVYQENVYVVEKKFQSF 385
Query: 404 QK-----GVPESNIIFLNLISAPKGVHVVC 428
+K G E++I ++L S KG + C
Sbjct: 386 KKVSRSMGYGENDISIVSLHSISKGYYGEC 475
>TC20082 weakly similar to UP|AAR27072 (AAR27072) EIN3-binding F-box protein
2, partial (9%)
Length = 630
Score = 26.6 bits (57), Expect = 9.5
Identities = 10/26 (38%), Positives = 17/26 (64%)
Frame = +1
Query: 421 PKGVHVVCKCFPRIKIVTSEIEIGLN 446
P V +VCKCFP + +V + ++ L+
Sbjct: 208 PVPVSLVCKCFPCLAVVIYQTKVWLS 285
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.141 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,070,656
Number of Sequences: 28460
Number of extensions: 91327
Number of successful extensions: 380
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 377
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 378
length of query: 476
length of database: 4,897,600
effective HSP length: 94
effective length of query: 382
effective length of database: 2,222,360
effective search space: 848941520
effective search space used: 848941520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC147712.11


