

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147537.13 + phase: 0
(127 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
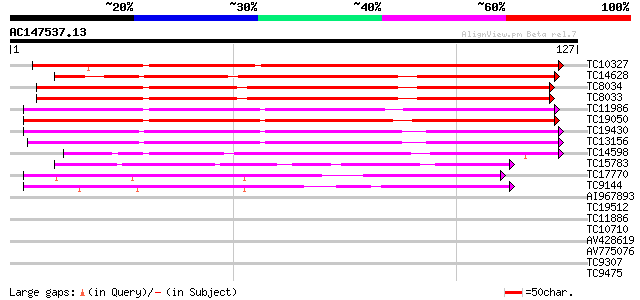
Score E
Sequences producing significant alignments: (bits) Value
TC10327 weakly similar to UP|NLT1_PRUDU (Q43017) Nonspecific lip... 174 5e-45
TC14628 similar to UP|NLTP_CICAR (O23758) Nonspecific lipid-tran... 108 2e-25
TC8034 similar to UP|NLTP_CICAR (O23758) Nonspecific lipid-trans... 105 2e-24
TC8033 similar to UP|NLTP_CICAR (O23758) Nonspecific lipid-trans... 105 2e-24
TC11986 weakly similar to UP|Q8RYA8 (Q8RYA8) Lipid transfer prec... 99 2e-22
TC19050 weakly similar to UP|Q9M6T9 (Q9M6T9) Nonspecific lipid-t... 95 3e-21
TC19430 weakly similar to UP|NLT3_HORVU (Q43766) Nonspecific lip... 88 4e-19
TC13156 weakly similar to UP|NLT3_HORVU (Q43766) Nonspecific lip... 86 1e-18
TC14598 65 5e-12
TC15783 similar to UP|Q9LJ86 (Q9LJ86) Gb|AAD48513.1 (AT3g22600/F... 48 6e-07
TC17770 weakly similar to UP|Q8VYI9 (Q8VYI9) AT5g64080/MHJ24_6, ... 43 1e-05
TC9144 39 4e-04
AI967893 37 0.001
TC19512 weakly similar to UP|O24101 (O24101) MtN5 protein precur... 30 0.095
TC11886 similar to UP|Q8XX59 (Q8XX59) Probable amino-acid-bindin... 28 0.47
TC10710 similar to UP|Q9SUZ2 (Q9SUZ2) 5B protein like protein, p... 27 1.1
AV428619 26 1.8
AV775076 26 2.4
TC9307 similar to PIR|T00914|T00914 leucine-rich repeat protein ... 25 3.1
TC9475 similar to UP|Q9FPW6 (Q9FPW6) POZ/BTB containing-protein ... 25 3.1
>TC10327 weakly similar to UP|NLT1_PRUDU (Q43017) Nonspecific lipid-transfer
protein 1 precursor (LTP 1), partial (52%)
Length = 683
Score = 174 bits (440), Expect = 5e-45
Identities = 91/123 (73%), Positives = 97/123 (77%), Gaps = 4/123 (3%)
Frame = +1
Query: 6 MATRLVCLAIV----CLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNG 61
M RLV LAIV VT P+AE AVT CG VV SLYPCVSY+MNGGN VP QCCNG
Sbjct: 79 MVARLVSLAIVFAVLVAVTTVPRAEGAVT-CGQVVNSLYPCVSYLMNGGNMVPG-QCCNG 252
Query: 62 IRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTD 121
IRNL MA T DRRAVCTCIK AVS SGFS+T+ NLNLAAGLP+KCGVNIPYQISPNTD
Sbjct: 253 IRNLYGMATNTPDRRAVCTCIKQAVSNSGFSFTSYNLNLAAGLPKKCGVNIPYQISPNTD 432
Query: 122 CNR 124
C+R
Sbjct: 433 CSR 441
>TC14628 similar to UP|NLTP_CICAR (O23758) Nonspecific lipid-transfer
protein precursor (LTP), partial (91%)
Length = 604
Score = 108 bits (271), Expect = 2e-25
Identities = 56/113 (49%), Positives = 72/113 (63%)
Frame = +1
Query: 11 VCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQ 70
+C+A+V P AEAA+ SCG V+ +L PC+ Y+ G P+ QCC G++NLN A
Sbjct: 115 ICMALVA----APMAEAAI-SCGSVIGALTPCMPYLTGG--PAPSPQCCAGVKNLNAAAS 273
Query: 71 TTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
TT DR+ C C+KNA S NLN AA LP KCGVNIPY+IS +T+CN
Sbjct: 274 TTPDRKTACGCLKNAAG----SMPNLNAGNAAALPGKCGVNIPYKISTSTNCN 420
>TC8034 similar to UP|NLTP_CICAR (O23758) Nonspecific lipid-transfer
protein precursor (LTP), partial (84%)
Length = 742
Score = 105 bits (263), Expect = 2e-24
Identities = 51/116 (43%), Positives = 71/116 (60%)
Frame = +1
Query: 7 ATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLN 66
+ + VC +V + P AEAA+T CG + ++L PC +YI G P+ CC G+R +N
Sbjct: 46 SVKFVCALVVLSMVVAPMAEAAIT-CGAIASALIPCTAYIQGGAG--PSPPCCAGVRRVN 216
Query: 67 TMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
+ A +T DRRA C C+K+A S+ N AA LP KCGVNIPY+IS +T+C
Sbjct: 217 SAASSTADRRAACNCLKSAAG----SFPRFNAGNAASLPGKCGVNIPYKISTSTNC 372
>TC8033 similar to UP|NLTP_CICAR (O23758) Nonspecific lipid-transfer
protein precursor (LTP), partial (85%)
Length = 707
Score = 105 bits (263), Expect = 2e-24
Identities = 51/116 (43%), Positives = 71/116 (60%)
Frame = +3
Query: 7 ATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLN 66
+ + VC +V + P AEAA+T CG + ++L PC +YI G P+ CC G+R +N
Sbjct: 78 SVKFVCALVVLSMVVAPMAEAAIT-CGAIASALIPCTAYIQGGAG--PSPPCCAGVRRVN 248
Query: 67 TMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
+ A +T DRRA C C+K+A S+ N AA LP KCGVNIPY+IS +T+C
Sbjct: 249 SAASSTADRRAACNCLKSAAG----SFPRFNAGNAASLPGKCGVNIPYKISTSTNC 404
>TC11986 weakly similar to UP|Q8RYA8 (Q8RYA8) Lipid transfer precursor
protein (Nonspecific lipid-transfer protein) (LTP),
partial (77%)
Length = 468
Score = 99.4 bits (246), Expect = 2e-22
Identities = 50/120 (41%), Positives = 72/120 (59%)
Frame = +2
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
+T+ + L ++C+V P AEAA+ CG V ++ PC+ Y+ + G +VPA CCNGIR
Sbjct: 74 TTLVLKFASLVVMCMVLGSPFAEAALP-CGQVQFTVAPCIGYLRSPGPSVPAP-CCNGIR 247
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
L A+T DR+ C C+K+ + S LNL A LP KCGVN Y+I+P+ +CN
Sbjct: 248 TLKNQAKTIADRQGACRCLKS----TSLSLPGLNLPALASLPGKCGVNFSYKITPSINCN 415
>TC19050 weakly similar to UP|Q9M6T9 (Q9M6T9) Nonspecific lipid-transfer
protein (LTP), partial (57%)
Length = 579
Score = 95.1 bits (235), Expect = 3e-21
Identities = 47/120 (39%), Positives = 73/120 (60%)
Frame = +2
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
+ + + LA++ +V P A+AA+ CG + ++ PC+ Y+ N TVPAA CCNGIR
Sbjct: 32 AALVLKFASLAVMVMVLGSPFADAALP-CGQLTFTVAPCIGYLRNPTPTVPAA-CCNGIR 205
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
++ A+T DR+ VC C+K+ V ++ +NL A +P KCG + Y+ISP+ DCN
Sbjct: 206 DILRQAKTVPDRQGVCRCLKSTV----YNLPGINLPALASVPGKCGAKLSYKISPSIDCN 373
>TC19430 weakly similar to UP|NLT3_HORVU (Q43766) Nonspecific lipid-transfer
protein 3 precursor (LTP 3) (CW20) (CW-20) (CW-19),
partial (53%)
Length = 520
Score = 88.2 bits (217), Expect = 4e-19
Identities = 44/121 (36%), Positives = 67/121 (55%)
Frame = +3
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
+TM A+V L+ +EAA+ SC V+ L PCVSY+++G P A CC+G +
Sbjct: 15 NTMKPSSAFSALVLLMLLVSASEAAI-SCSDVIKDLKPCVSYLVSGSGKPPGA-CCSGAK 188
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
L + T+ D++A C CIK+ +N LA LP CG+N+P +SP+ DC+
Sbjct: 189 ALASAVSTSEDKKAACNCIKSTAKS-----IKMNSQLAKALPGNCGINVPINVSPDADCS 353
Query: 124 R 124
+
Sbjct: 354 K 356
>TC13156 weakly similar to UP|NLT3_HORVU (Q43766) Nonspecific lipid-transfer
protein 3 precursor (LTP 3) (CW20) (CW-20) (CW-19),
partial (53%)
Length = 603
Score = 86.3 bits (212), Expect = 1e-18
Identities = 44/120 (36%), Positives = 65/120 (53%)
Frame = +1
Query: 5 TMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRN 64
TM A+V L+ +EAA+ SC V+ L PCVSY+++G P A CC+G +
Sbjct: 7 TMKLSSAFSALVLLMLLVSASEAAI-SCSDVIKDLKPCVSYLVSGSGKPPVA-CCSGAKA 180
Query: 65 LNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCNR 124
L + T+ D++A C CIK+ +N LA LP CG+N P +SP+ DC++
Sbjct: 181 LASAVSTSEDKKAACNCIKSTAKS-----IKMNSQLAEALPGNCGINTPISVSPDADCSK 345
>TC14598
Length = 613
Score = 64.7 bits (156), Expect = 5e-12
Identities = 37/113 (32%), Positives = 63/113 (55%), Gaps = 1/113 (0%)
Frame = +1
Query: 13 LAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTT 72
L IV L++ A+A +T C V+ ++ PC S+++ N+ P+ CC+G++ L+ A T
Sbjct: 79 LPIVILLSISA-AQATIT-CPAVIQAVTPCASFLLQ--NSSPSQGCCDGVKKLSGEAGTQ 246
Query: 73 NDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPY-QISPNTDCNR 124
+ A+C C+K ++Q G + N + LP+ CGV + I NTDC +
Sbjct: 247 EGKTAICQCLKQGLAQVG----KYDPNRVSELPKDCGVPLTLPPIDQNTDCTK 393
>TC15783 similar to UP|Q9LJ86 (Q9LJ86) Gb|AAD48513.1 (AT3g22600/F16J14_17),
partial (48%)
Length = 578
Score = 47.8 bits (112), Expect = 6e-07
Identities = 31/103 (30%), Positives = 54/103 (52%)
Frame = -2
Query: 11 VCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQ 70
+CL +V + T + AA + C +TSL PC++YI G ++ P+ CC+ L+++ Q
Sbjct: 421 LCLVMVMVATMWTQ-NAAQSGCTSALTSLSPCLNYI-TGSSSSPSPSCCS---QLSSVVQ 257
Query: 71 TTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIP 113
++ + +C+ + S G + +N LA LP C V P
Sbjct: 256 SS--PQCLCSLLNGGGSSFGIT---MNQTLALSLPGPCKVQTP 143
>TC17770 weakly similar to UP|Q8VYI9 (Q8VYI9) AT5g64080/MHJ24_6, partial
(42%)
Length = 554
Score = 43.1 bits (100), Expect = 1e-05
Identities = 30/117 (25%), Positives = 50/117 (42%), Gaps = 9/117 (7%)
Frame = +3
Query: 4 STMATR---LVCLAIVCLVTFGPKAE-----AAVTSCGPVVTSLYPCVSYIMNGGN-TVP 54
S MA++ ++C+ +C V F A A C ++ + C+S++ N T P
Sbjct: 168 SLMASKVSLILCVVAICAVDFAHSASSNNAPAPSADCSNLILKMTDCLSFVTNDSTVTKP 347
Query: 55 AAQCCNGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVN 111
QCC+G++++ A +C+ A S LN A LP C V+
Sbjct: 348 QGQCCSGLKSVLKTAP---------SCLCEAFKSSSQFGVVLNATKALALPSACKVS 491
>TC9144
Length = 835
Score = 38.5 bits (88), Expect = 4e-04
Identities = 32/124 (25%), Positives = 51/124 (40%), Gaps = 14/124 (11%)
Frame = +1
Query: 4 STMATRLVCLA--IVCLVTFGPKAEA-----------AVTSCGPVVTSLYPCVSYIMNGG 50
+T TR+V A ++ L G A++ A C +T++ C++++ G
Sbjct: 82 TTTTTRVVAFAALVLALACHGMSAQSLAPESASAPSPAGVDCFTALTNVSDCLTFVEAGS 261
Query: 51 N-TVPAAQCCNGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCG 109
N T P CC L + +C C + + F +NLN A LP CG
Sbjct: 262 NLTKPDKGCCPEFAGLI-------ESNPICLC--QLLGKPDFVGIKINLNKAIKLPSVCG 414
Query: 110 VNIP 113
V+ P
Sbjct: 415 VDTP 426
>AI967893
Length = 258
Score = 37.0 bits (84), Expect = 0.001
Identities = 20/55 (36%), Positives = 32/55 (57%)
Frame = +3
Query: 6 MATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCN 60
M LV +A++C AA +SC V+ SL PC++YI G ++ P++ CC+
Sbjct: 84 MGLILVVMAMLCA------GAAAQSSCTNVLVSLSPCLNYI-TGNSSTPSSGCCS 227
>TC19512 weakly similar to UP|O24101 (O24101) MtN5 protein precursor,
partial (52%)
Length = 430
Score = 30.4 bits (67), Expect = 0.095
Identities = 21/63 (33%), Positives = 27/63 (42%), Gaps = 1/63 (1%)
Frame = +2
Query: 54 PAAQCCNGIRNLNTMAQTTNDRRAVCTC-IKNAVSQSGFSYTNLNLNLAAGLPRKCGVNI 112
P +CC IR+ N C C K+ + G + TN A LPRKCG+
Sbjct: 29 PKEKCCAVIRHANL----------TCLCGYKSLLPSVGINPTN-----ALALPRKCGLKT 163
Query: 113 PYQ 115
P Q
Sbjct: 164 PRQ 172
>TC11886 similar to UP|Q8XX59 (Q8XX59) Probable amino-acid-binding
periplasmic ABC transporter protein, partial (5%)
Length = 504
Score = 28.1 bits (61), Expect = 0.47
Identities = 10/25 (40%), Positives = 16/25 (64%)
Frame = -3
Query: 44 SYIMNGGNTVPAAQCCNGIRNLNTM 68
SY +NGG+ + A CC+G L+ +
Sbjct: 241 SYTLNGGHLLIMASCCSGFTTLSCL 167
>TC10710 similar to UP|Q9SUZ2 (Q9SUZ2) 5B protein like protein, partial
(58%)
Length = 730
Score = 26.9 bits (58), Expect = 1.1
Identities = 18/73 (24%), Positives = 30/73 (40%)
Frame = +1
Query: 36 VTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTN 95
+ L PC + + P CCN +++L C C V +G +
Sbjct: 10 LVQLIPCRAAVAPFSPIPPNEACCNALKSLG----------QPCLC----VLVNGPPISG 147
Query: 96 LNLNLAAGLPRKC 108
++ N+A+ LP KC
Sbjct: 148 VDRNMASLLPEKC 186
>AV428619
Length = 405
Score = 26.2 bits (56), Expect = 1.8
Identities = 15/40 (37%), Positives = 19/40 (47%)
Frame = -2
Query: 63 RNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAA 102
R N +A ++ RAVC C VS L+LNL A
Sbjct: 257 RKTNNVASSSYSSRAVCNCCNEKVSLFPPLDLPLSLNLVA 138
>AV775076
Length = 469
Score = 25.8 bits (55), Expect = 2.4
Identities = 13/36 (36%), Positives = 19/36 (52%)
Frame = +3
Query: 34 PVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMA 69
P S C SY+ N+VPAAQ C+ + ++ A
Sbjct: 321 PYPRSRVLCSSYLPVSSNSVPAAQPCSWLEPVSLHA 428
>TC9307 similar to PIR|T00914|T00914 leucine-rich repeat protein F21B7.28 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(41%)
Length = 587
Score = 25.4 bits (54), Expect = 3.1
Identities = 9/27 (33%), Positives = 15/27 (55%)
Frame = -1
Query: 54 PAAQCCNGIRNLNTMAQTTNDRRAVCT 80
P CC+G + ++ A +DRR C+
Sbjct: 254 PRTGCCSGTNSAHSPASAISDRRIGCS 174
>TC9475 similar to UP|Q9FPW6 (Q9FPW6) POZ/BTB containing-protein AtPOB1,
partial (22%)
Length = 572
Score = 25.4 bits (54), Expect = 3.1
Identities = 14/52 (26%), Positives = 23/52 (43%), Gaps = 5/52 (9%)
Frame = +3
Query: 36 VTSLYPCVSYIMNGG-----NTVPAAQCCNGIRNLNTMAQTTNDRRAVCTCI 82
VT + C +++ G +TV CCN ++NLN + TC+
Sbjct: 372 VTRICICEPFLLVGSFDLFWDTVFLVTCCNNLKNLNLSLALHKTK*IFRTCL 527
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.134 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,583,077
Number of Sequences: 28460
Number of extensions: 33147
Number of successful extensions: 201
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 182
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 184
length of query: 127
length of database: 4,897,600
effective HSP length: 80
effective length of query: 47
effective length of database: 2,620,800
effective search space: 123177600
effective search space used: 123177600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 50 (23.9 bits)
Medicago: description of AC147537.13


