

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147517.5 + phase: 0
(167 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
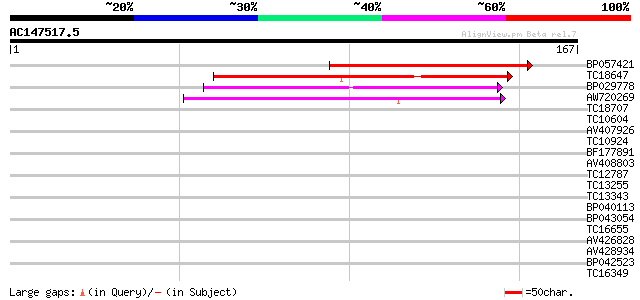
Score E
Sequences producing significant alignments: (bits) Value
BP057421 90 2e-19
TC18647 similar to UP|Q9ASR9 (Q9ASR9) At2g01320/F10A8.20, partia... 72 5e-14
BP029778 64 1e-11
AW720269 55 8e-09
TC18707 homologue to PIR|T02901|T02901 MSP1 protein homolog T13J... 37 0.001
TC10604 similar to AAQ89671 (AAQ89671) At4g33460, partial (36%) 35 0.009
AV407926 35 0.009
TC10924 homologue to UP|Q8H9D4 (Q8H9D4) 26S proteasome AAA-ATPas... 34 0.015
BF177891 33 0.025
AV408803 32 0.055
TC12787 weakly similar to UP|MSP1_YEAST (P28737) MSP1 protein (T... 31 0.095
TC13255 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (41%) 31 0.12
TC13343 similar to UP|Q9FMR9 (Q9FMR9) Ruv DNA-helicase-like prot... 31 0.12
BP040113 31 0.12
BP043054 30 0.21
TC16655 homologue to UP|O99018 (O99018) Chloroplast protease pre... 30 0.21
AV426828 30 0.21
AV428934 30 0.28
BP042523 30 0.28
TC16349 homologue to UP|PRS6_SOLTU (P54778) 26S protease regulat... 29 0.36
>BP057421
Length = 547
Score = 90.1 bits (222), Expect = 2e-19
Identities = 37/60 (61%), Positives = 53/60 (87%)
Frame = +3
Query: 95 KLDPKLKFSGRVTYNGHEMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQGVGPQY 154
K+D +L+ +G +TYNGH+++EFVP++TAAY+ QND+H+GE+TV+ETL FSAR QGVG +Y
Sbjct: 24 KMDHELRVTGEITYNGHKLNEFVPRKTAAYISQNDVHVGEMTVKETLDFSARCQGVGTRY 203
>TC18647 similar to UP|Q9ASR9 (Q9ASR9) At2g01320/F10A8.20, partial (17%)
Length = 430
Score = 72.0 bits (175), Expect = 5e-14
Identities = 42/90 (46%), Positives = 57/90 (62%), Gaps = 2/90 (2%)
Frame = +1
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKL--DPKLKFSGRVTYNGHEMSEFVP 118
+LK+VSG KP R+ ++GP SGKTTLL LAG+L P+L SG + +NG S+
Sbjct: 106 LLKNVSGEAKPGRLLAIMGPSGSGKTTLLNVLAGQLAASPRLHLSGLLEFNGKPGSKNAY 285
Query: 119 QRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
+ AYV Q DL +LTVRETL+ + +Q
Sbjct: 286 K--FAYVRQEDLFFSQLTVRETLSLATELQ 369
>BP029778
Length = 438
Score = 64.3 bits (155), Expect = 1e-11
Identities = 34/88 (38%), Positives = 52/88 (58%)
Frame = -2
Query: 58 RLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFV 117
+L +L +VSG+ P +T L+G +GKTTL+ LAG+ G + +G+ +
Sbjct: 287 KLKLLSNVSGVFAPGVLTALMGSSGAGKTTLMDVLAGRKTGGY-IEGDIKISGYPKVQHT 111
Query: 118 PQRTAAYVDQNDLHIGELTVRETLAFSA 145
R A YV+QND+H ++TV E+L FSA
Sbjct: 110 FARVAGYVEQNDIHSPQVTVEESLLFSA 27
>AW720269
Length = 524
Score = 54.7 bits (130), Expect = 8e-09
Identities = 33/96 (34%), Positives = 55/96 (56%), Gaps = 1/96 (1%)
Frame = +3
Query: 52 LPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGH 111
L + + +NILK VS + + S + ++GP +GK+TLL +AG++ + ++ N H
Sbjct: 27 LTQKPKPVNILKSVSFVARSSEIVAVVGPSGTGKSTLLRIIAGRVKDRDFDPKTISINDH 206
Query: 112 EM-SEFVPQRTAAYVDQNDLHIGELTVRETLAFSAR 146
M S ++ +V Q D + LTV+ETL FSA+
Sbjct: 207 PMTSPAQLRKICGFVAQEDNLLPLLTVKETLLFSAK 314
>TC18707 homologue to PIR|T02901|T02901 MSP1 protein homolog T13J8.110 -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(29%)
Length = 771
Score = 37.4 bits (85), Expect = 0.001
Identities = 22/54 (40%), Positives = 32/54 (58%)
Frame = +3
Query: 42 VESLLNSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGK 95
++ L L +LP R R ++ K G++KP R LL GPP +GKT L A+A +
Sbjct: 225 IKESLQELVMLPLR--RPDLFK--GGLLKPCRGILLFGPPGTGKTMLAXAIANE 374
>TC10604 similar to AAQ89671 (AAQ89671) At4g33460, partial (36%)
Length = 498
Score = 34.7 bits (78), Expect = 0.009
Identities = 26/61 (42%), Positives = 32/61 (51%)
Frame = +2
Query: 59 LNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVP 118
+ +L+D S I + +LLGP GK+TLL LAG L P SG V N E FV
Sbjct: 194 VRVLRDCSLRIPSGQFWMLLGPNGCGKSTLLKILAGLLAPT---SGTVYVN--EPKSFVF 358
Query: 119 Q 119
Q
Sbjct: 359 Q 361
>AV407926
Length = 309
Score = 34.7 bits (78), Expect = 0.009
Identities = 15/38 (39%), Positives = 25/38 (65%)
Frame = +1
Query: 94 GKLDPKLKFSGRVTYNGHEMSEFVPQRTAAYVDQNDLH 131
G L P L S ++ GH++S F+P+RT A ++Q +L+
Sbjct: 43 GHLTPYLHLSNKLAKRGHKISFFIPRRTQAKLEQFNLY 156
>TC10924 homologue to UP|Q8H9D4 (Q8H9D4) 26S proteasome AAA-ATPase subunit
RPT4a, partial (49%)
Length = 648
Score = 33.9 bits (76), Expect = 0.015
Identities = 15/29 (51%), Positives = 19/29 (64%)
Frame = +2
Query: 69 IKPSRMTLLLGPPSSGKTTLLLALAGKLD 97
IKP + LL GPP +GKT L A+A +D
Sbjct: 551 IKPPKGVLLYGPPGTGKTLLARAIASNID 637
>BF177891
Length = 390
Score = 33.1 bits (74), Expect = 0.025
Identities = 21/51 (41%), Positives = 28/51 (54%), Gaps = 4/51 (7%)
Frame = +1
Query: 47 NSLHVLPSRKQRL----NILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALA 93
+ L +LP R+ L N+L+ V GI LL GPP +GKT + ALA
Sbjct: 13 SELVILPMRRPELFTHGNLLRPVKGI-------LLFGPPGTGKTLMAKALA 144
>AV408803
Length = 305
Score = 32.0 bits (71), Expect = 0.055
Identities = 14/38 (36%), Positives = 22/38 (57%)
Frame = +3
Query: 94 GKLDPKLKFSGRVTYNGHEMSEFVPQRTAAYVDQNDLH 131
G L P L S ++ GH +S +P+RT A ++ +LH
Sbjct: 75 GHLTPFLHLSNKLARRGHRISFLIPKRTQAKLEHLNLH 188
>TC12787 weakly similar to UP|MSP1_YEAST (P28737) MSP1 protein
(TAT-binding homolog 4), partial (20%)
Length = 389
Score = 31.2 bits (69), Expect = 0.095
Identities = 19/51 (37%), Positives = 27/51 (52%)
Frame = +3
Query: 43 ESLLNSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALA 93
E L L +LP ++ L ++ KP + LL GPP +GKT L A+A
Sbjct: 3 EDTLKELVMLPLKRPELFCKGQLT---KPCKGILLFGPPGTGKTMLAKAVA 146
>TC13255 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (41%)
Length = 532
Score = 30.8 bits (68), Expect = 0.12
Identities = 14/31 (45%), Positives = 20/31 (64%)
Frame = +1
Query: 63 KDVSGIIKPSRMTLLLGPPSSGKTTLLLALA 93
K +G++ P + LL GPP +GKT L A+A
Sbjct: 7 KYFTGLLSPWKGILLFGPPGTGKTMLAKAVA 99
>TC13343 similar to UP|Q9FMR9 (Q9FMR9) Ruv DNA-helicase-like protein,
partial (21%)
Length = 518
Score = 30.8 bits (68), Expect = 0.12
Identities = 14/30 (46%), Positives = 18/30 (59%)
Frame = +1
Query: 73 RMTLLLGPPSSGKTTLLLALAGKLDPKLKF 102
R LL GPP +GKT L L + +L K+ F
Sbjct: 361 RALLLAGPPGTGKTALALGICQELGTKVPF 450
>BP040113
Length = 439
Score = 30.8 bits (68), Expect = 0.12
Identities = 14/30 (46%), Positives = 18/30 (59%)
Frame = +2
Query: 73 RMTLLLGPPSSGKTTLLLALAGKLDPKLKF 102
R LL GPP +GKT L L + +L K+ F
Sbjct: 281 RALLLAGPPGTGKTALALGICQELGTKVPF 370
>BP043054
Length = 467
Score = 30.0 bits (66), Expect = 0.21
Identities = 17/42 (40%), Positives = 27/42 (63%), Gaps = 1/42 (2%)
Frame = +1
Query: 70 KPS-RMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNG 110
KP+ R+ L++GP SGK++L+ A+A L + + GR T G
Sbjct: 217 KPAPRLNLVIGPNGSGKSSLVCAIALGLGGEPQLLGRATSIG 342
>TC16655 homologue to UP|O99018 (O99018) Chloroplast protease precursor,
partial (39%)
Length = 934
Score = 30.0 bits (66), Expect = 0.21
Identities = 17/46 (36%), Positives = 25/46 (53%), Gaps = 10/46 (21%)
Frame = +3
Query: 60 NILKDVSGIIKPSRMT----------LLLGPPSSGKTTLLLALAGK 95
+ ++ V + KP R T LL+GPP +GKT L A+AG+
Sbjct: 687 DFMEVVEFLKKPERFTAVGARIPKGVLLIGPPGTGKTLLAKAIAGE 824
>AV426828
Length = 401
Score = 30.0 bits (66), Expect = 0.21
Identities = 13/21 (61%), Positives = 17/21 (80%)
Frame = +1
Query: 76 LLLGPPSSGKTTLLLALAGKL 96
LL GPP +GKT+ +LA+A KL
Sbjct: 121 LLYGPPGTGKTSTILAVARKL 183
>AV428934
Length = 323
Score = 29.6 bits (65), Expect = 0.28
Identities = 14/41 (34%), Positives = 23/41 (55%)
Frame = +1
Query: 47 NSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTT 87
++L +PS +L+ + KP + +L+GPP SGK T
Sbjct: 58 SNLEDVPSVDLMTELLRRLKCSSKPDKRLILIGPPGSGKGT 180
>BP042523
Length = 479
Score = 29.6 bits (65), Expect = 0.28
Identities = 13/21 (61%), Positives = 17/21 (80%)
Frame = +1
Query: 76 LLLGPPSSGKTTLLLALAGKL 96
+L GPP +GKTT +LALA +L
Sbjct: 184 ILSGPPGTGKTTSILALAHEL 246
>TC16349 homologue to UP|PRS6_SOLTU (P54778) 26S protease regulatory subunit
6B homolog, partial (65%)
Length = 901
Score = 29.3 bits (64), Expect = 0.36
Identities = 14/25 (56%), Positives = 16/25 (64%)
Frame = +3
Query: 69 IKPSRMTLLLGPPSSGKTTLLLALA 93
I P R LL GPP +GKT L A+A
Sbjct: 591 IDPPRGVLLYGPPGTGKTMLAKAVA 665
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.138 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,495,665
Number of Sequences: 28460
Number of extensions: 29340
Number of successful extensions: 199
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 196
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 196
length of query: 167
length of database: 4,897,600
effective HSP length: 84
effective length of query: 83
effective length of database: 2,506,960
effective search space: 208077680
effective search space used: 208077680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC147517.5


