

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147496.3 - phase: 0
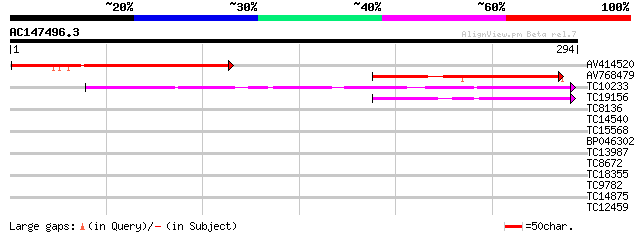
(294 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV414520 127 3e-30
AV768479 101 1e-22
TC10233 similar to UP|Q84ZV0 (Q84ZV0) R 14 protein, partial (32%) 64 3e-11
TC19156 similar to GB|AAL91278.1|19699336|AY090375 At2g15220/F15... 46 8e-06
TC8136 similar to GB|AAB60729.1|2160166|F21M12 F21M12.13 {Arabid... 28 1.8
TC14540 similar to UP|O49855 (O49855) Acid phosphatase , partial... 28 2.4
TC15568 28 2.4
BP046302 27 4.1
TC13987 similar to UP|Q45307 (Q45307) 5A2 divIB (Fragment), part... 26 7.0
TC8672 similar to UP|Q8SMH8 (Q8SMH8) RNA-binding protein, partia... 26 7.0
TC18355 weakly similar to UP|Q9SGD8 (Q9SGD8) T23G18.8, partial (... 26 9.1
TC9782 26 9.1
TC14875 similar to UP|Q9VNX6 (Q9VNX6) CG7421-PA, partial (4%) 26 9.1
TC12459 26 9.1
>AV414520
Length = 409
Score = 127 bits (318), Expect = 3e-30
Identities = 72/129 (55%), Positives = 94/129 (72%), Gaps = 14/129 (10%)
Frame = +1
Query: 2 ENEDKHFLFQPLL-PHFNPSS------YEI-----NQQND--YNNSKFFIPNNNNIIHRI 47
+ EDKH L QPL+ P PSS Y + + QND NN+K FI ++ +IIHR+
Sbjct: 22 KEEDKHSLLQPLIIPSLIPSSTTNTTLYSLCNPHHHHQNDPRNNNTKLFI-SDKSIIHRV 198
Query: 48 LLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVE 107
+L+L V+I+SIWAN+EASKTF I +VNDT D+ A RRF L YVSND+A+RILLNTSSFVE
Sbjct: 199 ILLLLVSILSIWANHEASKTFQITLVNDTNDTPAARRFALNYVSNDRAARILLNTSSFVE 378
Query: 108 QILYPNGNN 116
++LYP+ N+
Sbjct: 379 KLLYPHHND 405
>AV768479
Length = 462
Score = 101 bits (252), Expect = 1e-22
Identities = 54/107 (50%), Positives = 68/107 (63%), Gaps = 8/107 (7%)
Frame = -2
Query: 189 DGRSKASPRLLDGMVEYIAELAGFHRERLSGGVGESPECEDGREL-----WWDNKDPTHV 243
+G +A DGM EYI ++AGF R+ L PEC E WW++K+P HV
Sbjct: 461 NGGFRAPIGFFDGMAEYITKVAGFPRKGL-------PECGQKEEALGGLRWWEDKEPAHV 303
Query: 244 ARLLHYCEKY-DKGFIQRLNEAMKDTWHDRMVGDVLGLKATKL--CG 287
ARLLHYCE Y KGFI+RLNE M+DTWHDR+V +VLG+ T + CG
Sbjct: 302 ARLLHYCEDYYKKGFIRRLNETMRDTWHDRVVENVLGVPLTTMRPCG 162
>TC10233 similar to UP|Q84ZV0 (Q84ZV0) R 14 protein, partial (32%)
Length = 889
Score = 63.9 bits (154), Expect = 3e-11
Identities = 56/254 (22%), Positives = 106/254 (41%)
Frame = +1
Query: 40 NNNIIHRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRIL 99
N+ I+ + + F+ ++ + +K D + N + G RF + A + L
Sbjct: 7 NSPTINATMKMHFIGLLIFLTAMQGTKAVDYTVTNKALSTPGGVRFR-DQIGEASARQTL 183
Query: 100 LNTSSFVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINSYVIEI 159
+ + F+ +I N D +K ++ V+L + ++D + + +E ++++ +
Sbjct: 184 DSATQFIWRIFQQNTAAD------RKNVEKVSLFI--DDMDGVAYTSNNEIHLSARYVNS 339
Query: 160 SPMLLEDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGFHRERLSG 219
L+ E I G + M VW W+G A L++G+ +Y+ RL
Sbjct: 340 YGGDLKRE------ISGVLYHEMVHVWQWNGNGGAPGGLIEGIADYV---------RLKA 474
Query: 220 GVGESPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLG 279
S + G+ WD+ AR L YC GF+ LN+ M+ + D+ +LG
Sbjct: 475 DYAPSHWVKPGQGNKWDHGYDV-TARFLDYCNSLKNGFVAELNKKMRTGYSDQFFVQILG 651
Query: 280 LKATKLCGLYNATY 293
+L Y A Y
Sbjct: 652 KSVDQLWTDYKAKY 693
>TC19156 similar to GB|AAL91278.1|19699336|AY090375 At2g15220/F15A23.4
{Arabidopsis thaliana;}, partial (41%)
Length = 479
Score = 45.8 bits (107), Expect = 8e-06
Identities = 31/105 (29%), Positives = 47/105 (44%)
Frame = +1
Query: 189 DGRSKASPRLLDGMVEYIAELAGFHRERLSGGVGESPECEDGRELWWDNKDPTHVARLLH 248
DG +A L++G+ +++ AG+ L G G + W D T AR L
Sbjct: 1 DGNGQAPSGLIEGIADFVRLKAGYAPLDLWVPPG-------GGDKWDQGYDVT--ARFLD 153
Query: 249 YCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKATKLCGLYNATY 293
YC GF+ LN+ MK ++ + +LG T+L Y A Y
Sbjct: 154 YCNSIRNGFVGELNKKMKSSYSETYFTQLLGKSVTQLWTGYKANY 288
>TC8136 similar to GB|AAB60729.1|2160166|F21M12 F21M12.13 {Arabidopsis
thaliana;}, partial (70%)
Length = 1659
Score = 28.1 bits (61), Expect = 1.8
Identities = 14/44 (31%), Positives = 22/44 (49%)
Frame = -2
Query: 74 NDTKDSLAGRRFTLFYVSNDKASRILLNTSSFVEQILYPNGNND 117
N KD+ F +F+ + KAS + T SF +PN ++D
Sbjct: 1565 NIQKDNRINFTFNIFFPARHKASLFTVATPSFFNTTKHPNWDSD 1434
>TC14540 similar to UP|O49855 (O49855) Acid phosphatase , partial (88%)
Length = 1144
Score = 27.7 bits (60), Expect = 2.4
Identities = 16/51 (31%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Frame = +3
Query: 229 DGRELWWDNKDPTHVARLLHYCEKYDKGF-IQRLNEAMKDTWHDRMVGDVL 278
DG+ +W + D T ++ L +Y E GF ++ N+ + W DR VL
Sbjct: 354 DGKNIWVFDIDETSLSNLPYYAE---HGFGLELYNDTAFNLWVDRAAAPVL 497
>TC15568
Length = 637
Score = 27.7 bits (60), Expect = 2.4
Identities = 12/50 (24%), Positives = 26/50 (52%)
Frame = -2
Query: 40 NNNIIHRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFY 89
NN II+ +++L ++ S W Y ++T + + + +++ LFY
Sbjct: 603 NNRIINSRIILLIMSFYSTWIRYP*TRTSYYSLAYRSYTNCNNKQYILFY 454
>BP046302
Length = 435
Score = 26.9 bits (58), Expect = 4.1
Identities = 13/23 (56%), Positives = 15/23 (64%), Gaps = 3/23 (13%)
Frame = -2
Query: 7 HFLFQPLLPHFNP---SSYEINQ 26
H L PLLPHF+P SSY +Q
Sbjct: 413 HLLSHPLLPHFHPHFHSSYHHHQ 345
>TC13987 similar to UP|Q45307 (Q45307) 5A2 divIB (Fragment), partial (8%)
Length = 491
Score = 26.2 bits (56), Expect = 7.0
Identities = 9/15 (60%), Positives = 11/15 (73%)
Frame = +2
Query: 234 WWDNKDPTHVARLLH 248
+W N PTHV RL+H
Sbjct: 101 YWSNLMPTHVRRLIH 145
>TC8672 similar to UP|Q8SMH8 (Q8SMH8) RNA-binding protein, partial (26%)
Length = 665
Score = 26.2 bits (56), Expect = 7.0
Identities = 17/49 (34%), Positives = 23/49 (46%)
Frame = +1
Query: 15 PHFNPSSYEINQQNDYNNSKFFIPNNNNIIHRILLILFVAIISIWANYE 63
P + S + Q +NS F P+ + HR L+ FVA S WA E
Sbjct: 217 PILSNSLKTLKLQLPCSNSSPFSPSLLSTTHRSPLLTFVAQTSDWAQQE 363
>TC18355 weakly similar to UP|Q9SGD8 (Q9SGD8) T23G18.8, partial (19%)
Length = 575
Score = 25.8 bits (55), Expect = 9.1
Identities = 16/35 (45%), Positives = 20/35 (56%), Gaps = 4/35 (11%)
Frame = +2
Query: 126 IIKSVTL----RLARQNLDTITTITADEKNINSYV 156
II+S TL L + TITT+ ADE+ I YV
Sbjct: 443 IIRSFTLDLRLSLTMPSFVTITTLCADEEGITFYV 547
>TC9782
Length = 798
Score = 25.8 bits (55), Expect = 9.1
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = -3
Query: 37 IPNNNNIIHRILLILFVAIISI 58
IP N NI+H I LI F + I
Sbjct: 601 IPTNYNILHSIFLISFTGVSGI 536
>TC14875 similar to UP|Q9VNX6 (Q9VNX6) CG7421-PA, partial (4%)
Length = 1240
Score = 25.8 bits (55), Expect = 9.1
Identities = 13/39 (33%), Positives = 21/39 (53%)
Frame = -1
Query: 6 KHFLFQPLLPHFNPSSYEINQQNDYNNSKFFIPNNNNII 44
++FL L N + +I QQND+NN+ N+I+
Sbjct: 1039 RYFLVTILQAERNQNK*QIQQQNDHNNTPLIQACFNSIV 923
>TC12459
Length = 797
Score = 25.8 bits (55), Expect = 9.1
Identities = 10/24 (41%), Positives = 18/24 (74%)
Frame = +2
Query: 35 FFIPNNNNIIHRILLILFVAIISI 58
F + NN+NII I++++FV + S+
Sbjct: 524 FLVDNNHNIIIIIIIMVFVILYSL 595
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,016,920
Number of Sequences: 28460
Number of extensions: 68528
Number of successful extensions: 380
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 372
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 375
length of query: 294
length of database: 4,897,600
effective HSP length: 90
effective length of query: 204
effective length of database: 2,336,200
effective search space: 476584800
effective search space used: 476584800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC147496.3


