

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147484.10 - phase: 0 /pseudo
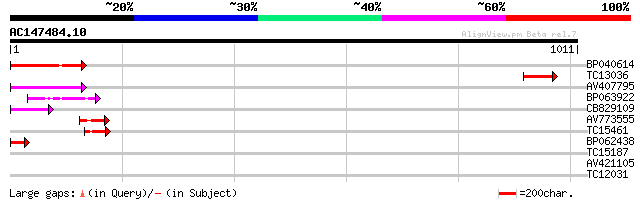
(1011 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP040614 97 1e-20
TC13036 similar to UP|Q9LDN0 (Q9LDN0) Phragmoplast-associated ki... 67 2e-11
AV407795 65 4e-11
BP063922 49 5e-06
CB829109 47 1e-05
AV773555 45 4e-05
TC15461 similar to UP|O24147 (O24147) Kinesin-like protein, part... 45 4e-05
BP062438 45 8e-05
TC15187 similar to UP|O24147 (O24147) Kinesin-like protein, part... 40 0.002
AV421105 40 0.002
TC12031 homologue to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-l... 36 0.035
>BP040614
Length = 504
Score = 97.1 bits (240), Expect = 1e-20
Identities = 64/140 (45%), Positives = 89/140 (62%), Gaps = 3/140 (2%)
Frame = +1
Query: 1 IYNEQITDLL--DPSQRNLQIREDVK-SGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIG 57
IYNEQ+ DLL + S R L IR + + +G+ V + V+ +DV L+ G NR +G
Sbjct: 97 IYNEQVRDLLVSNGSNRRLDIRNNSQLNGLNVPDAYLVPVTCTQDVLYLMEVGQKNRAVG 276
Query: 58 ATSINSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLK 117
AT++N SSRSH+V T V R + + + R ++LVDLAGSER + + A GERLK
Sbjct: 277 ATALNERSSRSHSVLTVHVRGR-ELVSGSILR---GCLHLVDLAGSERVEKSEAVGERLK 444
Query: 118 EAGNINRSLSQLGNLINILA 137
EA +INRSLS LG++I+ LA
Sbjct: 445 EAQHINRSLSALGDVISALA 504
>TC13036 similar to UP|Q9LDN0 (Q9LDN0) Phragmoplast-associated
kinesin-related protein 1, partial (4%)
Length = 480
Score = 66.6 bits (161), Expect = 2e-11
Identities = 43/62 (69%), Positives = 47/62 (75%), Gaps = 1/62 (1%)
Frame = +3
Query: 916 GYGCRAESCKGLPTN**VEEKT*EGGCIT**ACFRSTFAEEIDTTSL-**CCHAQL***L 974
GYGCRA SC GL T * V+E+T*EG +T**A RSTFAE I TT L **C HA +***L
Sbjct: 3 GYGCRAGSC*GLRTR*QVKEET*EGDYLT**ASGRSTFAEGISTTDL***CGHAHI***L 182
Query: 975 KG 976
KG
Sbjct: 183 KG 188
>AV407795
Length = 424
Score = 65.5 bits (158), Expect = 4e-11
Identities = 45/137 (32%), Positives = 70/137 (50%), Gaps = 1/137 (0%)
Frame = +2
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IY ++ DLL+ ++ L IRED K V + L E VS ++ + +L+ +G + R G T
Sbjct: 11 IYGGKLFDLLN-DRKKLFIREDGKQQVCIVGLQEYCVSDVESIKELIERGNATRSTGTTG 187
Query: 61 INSESSRSHTVFTCVVESRCKSAADGVSRFKTSRINLVDLAGSERQKSTGAAGERLK-EA 119
N ESSRSH + ++ R + +++ +DLAGSER T ++ + E
Sbjct: 188 ANEESSRSHAILQLAIK-RSVDGNESKPPRPVGKLSFIDLAGSERGADTTDNDKQTRIEG 364
Query: 120 GNINRSLSQLGNLINIL 136
IN+SL L I L
Sbjct: 365 AEINKSLLALKECIRAL 415
>BP063922
Length = 537
Score = 48.5 bits (114), Expect = 5e-06
Identities = 39/131 (29%), Positives = 68/131 (51%)
Frame = +2
Query: 32 LTEEQVSTMKDVTQLLLKGLSNRRIGATSINSESSRSHTVFTCVVESRCKSAADGVSRFK 91
L +E+V D + +L +R IN SH + T + + G + +
Sbjct: 182 LAQEKVDNPLDFSTVLKAAFQSRGNDLLKINV----SHLIVT--IHIFYNNLITGENSY- 340
Query: 92 TSRINLVDLAGSERQKSTGAAGERLKEAGNINRSLSQLGNLINILAEVSQTGKQRHIPYR 151
S+++LVDLAGSE + +GER+ + ++ +SLS LG++++ S T K+ +PY
Sbjct: 341 -SKLSLVDLAGSEGSITEDDSGERVTDLLHVMKSLSALGDVLS-----SLTSKKDIVPYE 502
Query: 152 DSRLTFLLQES 162
+S LT +L +S
Sbjct: 503 NSALTKVLADS 535
>CB829109
Length = 514
Score = 47.4 bits (111), Expect = 1e-05
Identities = 28/77 (36%), Positives = 44/77 (56%)
Frame = +2
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEEQVSTMKDVTQLLLKGLSNRRIGATS 60
IY ++ DLL ++ L +RED + V + L E +V ++ V + + KG + R G+T
Sbjct: 200 IYGGKLFDLLS-DRKKLCMREDGRQQVCIVGLQEFEVFDVQIVKEFIEKGNAARSTGSTG 376
Query: 61 INSESSRSHTVFTCVVE 77
N ESSRSH + VV+
Sbjct: 377 ANEESSRSHAILQLVVK 427
>AV773555
Length = 460
Score = 45.4 bits (106), Expect = 4e-05
Identities = 25/53 (47%), Positives = 35/53 (65%)
Frame = -1
Query: 125 SLSQLGNLINILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISP 177
SLS L ++I LA+ + HIP+R S+LT+LLQ +LGG++K M ISP
Sbjct: 460 SLSSLSDVIFALAQ-----QDDHIPFRHSKLTYLLQPALGGDSKTVMFVNISP 317
>TC15461 similar to UP|O24147 (O24147) Kinesin-like protein, partial (9%)
Length = 527
Score = 45.4 bits (106), Expect = 4e-05
Identities = 21/47 (44%), Positives = 33/47 (69%)
Frame = +3
Query: 134 NILAEVSQTGKQRHIPYRDSRLTFLLQESLGGNAKLAMVCAISPAQS 180
++++ +S G+ HIPYR+ +LT L+ +SLGGNAK M +SP +S
Sbjct: 6 DVISALSSGGQ--HIPYRNHKLTMLMSDSLGGNAKTLMFVNVSPIES 140
>BP062438
Length = 498
Score = 44.7 bits (104), Expect = 8e-05
Identities = 20/35 (57%), Positives = 25/35 (71%)
Frame = +3
Query: 1 IYNEQITDLLDPSQRNLQIREDVKSGVYVENLTEE 35
IYNE+I DLL P R LQI E+++ G+YV L EE
Sbjct: 393 IYNEEINDLLAPEHRKLQIHENLERGIYVAGLREE 497
>TC15187 similar to UP|O24147 (O24147) Kinesin-like protein, partial (8%)
Length = 611
Score = 40.0 bits (92), Expect = 0.002
Identities = 17/33 (51%), Positives = 24/33 (72%)
Frame = +2
Query: 148 IPYRDSRLTFLLQESLGGNAKLAMVCAISPAQS 180
IPYR+ +LT L+ +SLGGNA+ M +SP +S
Sbjct: 2 IPYRNHKLTMLMSDSLGGNAQTLMFVNVSPVES 100
>AV421105
Length = 415
Score = 39.7 bits (91), Expect = 0.002
Identities = 15/32 (46%), Positives = 25/32 (77%)
Frame = +2
Query: 148 IPYRDSRLTFLLQESLGGNAKLAMVCAISPAQ 179
+PYR+S+LT +LQ+SLGG ++ MV ++P +
Sbjct: 5 VPYRESKLTRILQDSLGGTSRALMVACLNPGE 100
>TC12031 homologue to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-like protein
A), partial (7%)
Length = 740
Score = 35.8 bits (81), Expect = 0.035
Identities = 18/32 (56%), Positives = 23/32 (71%)
Frame = +2
Query: 151 RDSRLTFLLQESLGGNAKLAMVCAISPAQSLV 182
R+S+LT+LLQ LGG++K M ISP QS V
Sbjct: 2 RNSKLTYLLQPCLGGDSKTLMFVNISPDQSSV 97
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.375 0.168 0.667
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,313,933
Number of Sequences: 28460
Number of extensions: 223621
Number of successful extensions: 3334
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 1287
Number of HSP's successfully gapped in prelim test: 110
Number of HSP's that attempted gapping in prelim test: 2002
Number of HSP's gapped (non-prelim): 1507
length of query: 1011
length of database: 4,897,600
effective HSP length: 99
effective length of query: 912
effective length of database: 2,080,060
effective search space: 1897014720
effective search space used: 1897014720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 13 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 35 (21.5 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147484.10


