

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147483.4 + phase: 0
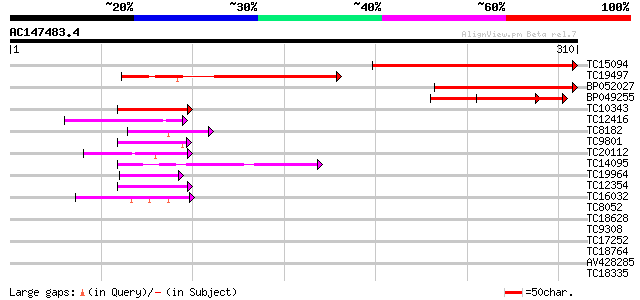
(310 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15094 similar to UP|Q8LA02 (Q8LA02) Contains similarity to RNA... 203 3e-53
TC19497 weakly similar to UP|Q8LA02 (Q8LA02) Contains similarity... 132 1e-31
BP052027 127 2e-30
BP049255 59 1e-09
TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (... 52 1e-07
TC12416 similar to GB|AAH56066.1|33417132|BC056066 sop-prov prot... 49 1e-06
TC8182 weakly similar to UP|Q93VA8 (Q93VA8) At1g76010/T4O12_22, ... 43 6e-05
TC9801 homologue to UP|Q8LAC6 (Q8LAC6) Glycine rich protein-like... 42 1e-04
TC20112 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich... 40 4e-04
TC14095 homologue to UP|RS2_ARATH (P49688) 40S ribosomal protein... 40 7e-04
TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%) 40 7e-04
TC12354 similar to UP|O23702 (O23702) Dehydrogenase, partial (25%) 40 7e-04
TC16032 similar to BAC87886 (BAC87886) PGL-3, partial (4%) 39 9e-04
TC8052 similar to GB|AAP13431.1|30023796|BT006323 At5g02530 {Ara... 38 0.002
TC18628 weakly similar to PIR|T09527|T09527 glycine-rich protein... 38 0.002
TC9308 weakly similar to UP|Q9SQ55 (Q9SQ55) Nuclear RNA binding ... 37 0.003
TC17252 similar to UP|O04237 (O04237) Transcription factor, part... 37 0.004
TC18764 similar to UP|Q8LBJ5 (Q8LBJ5) Nucleic acid binding prote... 37 0.004
AV428285 37 0.004
TC18335 similar to UP|Q7XJ13 (Q7XJ13) Chloroplast nucleoid DNA-b... 37 0.006
>TC15094 similar to UP|Q8LA02 (Q8LA02) Contains similarity to RNA-binding
protein from Arabidopsis thaliana gi|2129727 and
contains RNA recognition PF|00076 domain, partial (21%)
Length = 599
Score = 203 bits (516), Expect = 3e-53
Identities = 97/112 (86%), Positives = 105/112 (93%)
Frame = +2
Query: 199 RDALEKMKPFLMTYEGIRSQEEWEEVIEELMQRVPLLKKIVDHYSGPDRVTAKKQQEELE 258
RDALEKMKPFLM YEGI+SQEEWEE +EE M RVPL+KKIVDHY GPDRVTAKKQQ ELE
Sbjct: 2 RDALEKMKPFLMVYEGIQSQEEWEEAMEETMARVPLMKKIVDHYCGPDRVTAKKQQGELE 181
Query: 259 RVAKTLPTSAPSSVKEFTNRAVVSLQSNPGWGFDKKCQFMDKLVFEVSQHHK 310
RVAKTLPTSAPSSVK+FT+RAV+SLQSNPGWGFDKKC FMDKLV+EVSQH+K
Sbjct: 182 RVAKTLPTSAPSSVKQFTDRAVISLQSNPGWGFDKKCHFMDKLVWEVSQHYK 337
>TC19497 weakly similar to UP|Q8LA02 (Q8LA02) Contains similarity to
RNA-binding protein from Arabidopsis thaliana gi|2129727
and contains RNA recognition PF|00076 domain, partial
(11%)
Length = 436
Score = 132 bits (331), Expect = 1e-31
Identities = 74/122 (60%), Positives = 84/122 (68%), Gaps = 2/122 (1%)
Frame = +2
Query: 62 DGDGSGRGRGRGRGRDVYARGRGDRGRGG--RGGRGDGRGGFKRYGDDRTSHQDIARSNA 119
+G G GRGRGRGRGR RGRG RGR G R GR QD S+A
Sbjct: 113 EGGGLGRGRGRGRGR---GRGRGFRGRDGEERSGRS----------------QDTDISHA 235
Query: 120 DGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDINCAIEFEP 179
GLY+GD+ADGEKLAKKLGPEIM+Q+TE +EE+ RVLPSPL+DE VEAMD+N AIE EP
Sbjct: 236 GGLYLGDDADGEKLAKKLGPEIMNQLTEGFEEMASRVLPSPLEDELVEAMDVNYAIECEP 415
Query: 180 EY 181
EY
Sbjct: 416 EY 421
>BP052027
Length = 472
Score = 127 bits (319), Expect = 2e-30
Identities = 61/78 (78%), Positives = 69/78 (88%)
Frame = -1
Query: 233 PLLKKIVDHYSGPDRVTAKKQQEELERVAKTLPTSAPSSVKEFTNRAVVSLQSNPGWGFD 292
PL+KKIVDHY GPDRVT KK ELERVAKTLP SAPSSVK FT+RAV+SLQS+PGWGFD
Sbjct: 472 PLMKKIVDHYCGPDRVTPKKPPGELERVAKTLPPSAPSSVKPFTDRAVISLQSHPGWGFD 293
Query: 293 KKCQFMDKLVFEVSQHHK 310
+KC FMDKLV+EVSQ ++
Sbjct: 292 QKCPFMDKLVWEVSQPYQ 239
>BP049255
Length = 468
Score = 58.5 bits (140), Expect = 1e-09
Identities = 33/61 (54%), Positives = 38/61 (62%), Gaps = 1/61 (1%)
Frame = -1
Query: 231 RVPLLKKIVDHYSGPDRVTAKKQQEELER-VAKTLPTSAPSSVKEFTNRAVVSLQSNPGW 289
RVPL+KKIVDHY GPDRVTAKKQQ E R + K P + +FT+RAV P
Sbjct: 465 RVPLMKKIVDHYCGPDRVTAKKQQREN*RELLKLFPQALHLP*SQFTDRAVYLSSEQPRL 286
Query: 290 G 290
G
Sbjct: 285 G 283
Score = 47.8 bits (112), Expect = 2e-06
Identities = 26/51 (50%), Positives = 34/51 (65%), Gaps = 1/51 (1%)
Frame = -3
Query: 256 ELERVAKTLPTSAPSSVKE-FTNRAVVSLQSNPGWGFDKKCQFMDKLVFEV 305
ELERVAKTLPTSAPSS+K + R + ++ ++KC FMDKL+ V
Sbjct: 388 ELERVAKTLPTSAPSSLKPIY*PRCLSLFRATQAGELNQKCHFMDKLILGV 236
>TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (85%)
Length = 1116
Score = 52.4 bits (124), Expect = 1e-07
Identities = 26/41 (63%), Positives = 26/41 (63%)
Frame = +1
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
R G G G GRG GRG ARG G G GGRGGRG GRGG
Sbjct: 88 RGRGRGDGGGRGGGRGTPFKARGGGRGGGGGRGGRGGGRGG 210
Score = 38.9 bits (89), Expect = 0.001
Identities = 26/44 (59%), Positives = 26/44 (59%), Gaps = 3/44 (6%)
Frame = +1
Query: 68 RGRGRGRGRDVYARGRGDRGRG--GRGGRGDGRG-GFKRYGDDR 108
RGRG G G RGRGDRGRG GGRG GRG FK G R
Sbjct: 46 RGRGGGGG----FRGRGDRGRGRGDGGGRGGGRGTPFKARGGGR 165
Score = 35.8 bits (81), Expect = 0.009
Identities = 24/41 (58%), Positives = 24/41 (58%), Gaps = 7/41 (17%)
Frame = +1
Query: 67 GRGRGRG-RGRDVYARGRGDRG--RGGRG----GRGDGRGG 100
GRG G G RGR RGRGD G GGRG RG GRGG
Sbjct: 49 GRGGGGGFRGRGDRGRGRGDGGGRGGGRGTPFKARGGGRGG 171
>TC12416 similar to GB|AAH56066.1|33417132|BC056066 sop-prov protein
{Xenopus laevis;} , partial (32%)
Length = 416
Score = 48.5 bits (114), Expect = 1e-06
Identities = 31/68 (45%), Positives = 36/68 (52%), Gaps = 1/68 (1%)
Frame = +3
Query: 31 VNRHIRVRQTPADAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGR-DVYARGRGDRGRG 89
+NRH+ + T + R GDG GRGRG GRGR + RGRG RGRG
Sbjct: 33 INRHLLCQPTIN*LKMSGDGRGASGEPSAEGFGDGFGRGRGEGRGRGEGRGRGRG-RGRG 209
Query: 90 GRGGRGDG 97
RG RGDG
Sbjct: 210RRGRRGDG 233
Score = 33.9 bits (76), Expect = 0.036
Identities = 34/115 (29%), Positives = 47/115 (40%), Gaps = 16/115 (13%)
Frame = +3
Query: 69 GRGRGRGRDVYARGRGD---RGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADGLYVG 125
G GRG + A G GD RGRG GRG+GRG + G R + G
Sbjct: 84 GDGRGASGEPSAEGFGDGFGRGRGEGRGRGEGRGRGRGRGRGRRGRR------------G 227
Query: 126 DNADGE-----KLAKKLGPEIMDQITEAYE--------EIIERVLPSPLQDEYVE 167
D E KL + + ++ + E Y EII+ L L+DE ++
Sbjct: 228 DGDKKEWVPVTKLGRLVKAGMIKSLEEIYVFSLAIKEFEIIDHFLGDSLKDEVMK 392
>TC8182 weakly similar to UP|Q93VA8 (Q93VA8) At1g76010/T4O12_22, partial
(13%)
Length = 611
Score = 43.1 bits (100), Expect = 6e-05
Identities = 30/68 (44%), Positives = 31/68 (45%), Gaps = 21/68 (30%)
Frame = +3
Query: 65 GSGRGRGRGRGRDVYARGRGD--------------------RGRG-GRGGRGDGRGGFKR 103
G GRG GRGRGR RGRG RGRG GRGGRG GRG
Sbjct: 81 GRGRGFGRGRGRGFRGRGRGGYGAQPVGYYDYGEYDAPPAPRGRGRGRGGRGRGRGRNAS 260
Query: 104 YGDDRTSH 111
+G SH
Sbjct: 261 WGVAA*SH 284
Score = 35.8 bits (81), Expect = 0.009
Identities = 27/63 (42%), Positives = 30/63 (46%), Gaps = 1/63 (1%)
Frame = +3
Query: 58 FVRDDGDGSGRGRGRGRGRDVYARGRG-DRGRGGRGGRGDGRGGFKRYGDDRTSHQDIAR 116
F + G G G G GR RGRG RGRG RG RG GRGG YG + D
Sbjct: 15 FYNNGGMEYGGGDGWDGGRGYGGRGRGFGRGRG-RGFRGRGRGG---YGAQPVGYYDYGE 182
Query: 117 SNA 119
+A
Sbjct: 183 YDA 191
Score = 28.5 bits (62), Expect = 1.5
Identities = 15/38 (39%), Positives = 18/38 (46%)
Frame = +3
Query: 53 QPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGG 90
QP+ + + D RGRGRGR RGRG G
Sbjct: 153 QPVGYYDYGEYDAPPAPRGRGRGRGGRGRGRGRNASWG 266
>TC9801 homologue to UP|Q8LAC6 (Q8LAC6) Glycine rich protein-like, complete
Length = 981
Score = 42.4 bits (98), Expect = 1e-04
Identities = 23/45 (51%), Positives = 27/45 (59%), Gaps = 5/45 (11%)
Frame = +2
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGG-----RGDGRG 99
R++G G +G GRG D A+G G RGRGG GG RG GRG
Sbjct: 404 REEGAGGRPAKGPGRGFDDGAKGPGGRGRGGLGGKPGGSRGAGRG 538
>TC20112 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich
glycoprotein DZ-HRGP precursor, partial (11%)
Length = 523
Score = 40.4 bits (93), Expect = 4e-04
Identities = 28/62 (45%), Positives = 30/62 (48%), Gaps = 2/62 (3%)
Frame = +2
Query: 41 PADAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDV--YARGRGDRGRGGRGGRGDGR 98
PA + N P P F G G G GRG R D Y+ G G RGRGG RG GR
Sbjct: 299 PAAHANPNPPFVPPSGGFSVGRGGGFG-GRGGDRKYDTGRYSGGGGGRGRGGGSIRGGGR 475
Query: 99 GG 100
GG
Sbjct: 476 GG 481
Score = 36.6 bits (83), Expect = 0.006
Identities = 23/40 (57%), Positives = 23/40 (57%)
Frame = +2
Query: 61 DDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
D G SG G GRGRG G RG GGRGGRG G GG
Sbjct: 404 DTGRYSGGGGGRGRG------GGSIRG-GGRGGRGGGFGG 502
Score = 26.2 bits (56), Expect = 7.5
Identities = 15/41 (36%), Positives = 17/41 (40%)
Frame = +2
Query: 90 GRGGRGDGRGGFKRYGDDRTSHQDIARSNADGLYVGDNADG 130
GRGG GRGG ++Y R S R G G G
Sbjct: 359 GRGGGFGGRGGDRKYDTGRYSGGGGGRGRGGGSIRGGGRGG 481
>TC14095 homologue to UP|RS2_ARATH (P49688) 40S ribosomal protein S2,
partial (88%)
Length = 1134
Score = 39.7 bits (91), Expect = 7e-04
Identities = 34/112 (30%), Positives = 47/112 (41%)
Frame = +3
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNA 119
R GD G GRG G GRGDRGRG R GRGG +R G R +
Sbjct: 63 RGGGDRGGFGRGFG--------GRGDRGRGDR-----GRGGRRRTGGRRDEEEKWVPVTK 203
Query: 120 DGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDI 171
G V D + L + + +II+ ++ L+DE ++ M +
Sbjct: 204 LGRLVRDGK-----IRSLEQIYLHSLPIKEHQIIDMLVGPTLKDEVMKIMPV 344
>TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%)
Length = 562
Score = 39.7 bits (91), Expect = 7e-04
Identities = 20/35 (57%), Positives = 20/35 (57%)
Frame = +2
Query: 61 DDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRG 95
D G G GRGR GR GRG G GGRGGRG
Sbjct: 194 DSGRGGGRGRFGSGGRGGDRGGRGGSGGGGRGGRG 298
Score = 36.6 bits (83), Expect = 0.006
Identities = 22/41 (53%), Positives = 22/41 (53%)
Frame = +2
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
R G SG GRG GR GRG G GGRGG GRGG
Sbjct: 149 RSGGGRSGGGRG-GRFDSGRGGGRGRFGSGGRGGDRGGRGG 268
Score = 35.4 bits (80), Expect = 0.012
Identities = 26/47 (55%), Positives = 26/47 (55%), Gaps = 6/47 (12%)
Frame = +2
Query: 60 RDDGDGSGR---GRGRGRGRDVYARGRGDRG--RGGRGGR-GDGRGG 100
R G GR GRG GRGR G G RG RGGRGG G GRGG
Sbjct: 164 RSGGGRGGRFDSGRGGGRGRF----GSGGRGGDRGGRGGSGGGGRGG 292
Score = 30.8 bits (68), Expect = 0.31
Identities = 22/47 (46%), Positives = 22/47 (46%)
Frame = +2
Query: 62 DGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
D GSG G GR GRG R GRGG G GR G G DR
Sbjct: 128 DNQGSGGRSGGGRS----GGGRGGRFDSGRGG-GRGRFGSGGRGGDR 253
>TC12354 similar to UP|O23702 (O23702) Dehydrogenase, partial (25%)
Length = 488
Score = 39.7 bits (91), Expect = 7e-04
Identities = 20/41 (48%), Positives = 23/41 (55%)
Frame = -1
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
R G G RG+ R RGR RGRG +G G G R +GR G
Sbjct: 281 RGGGRGQQRGKSRRRGRRSRGRGRGGKGGDGAGRRVEGRRG 159
Score = 30.8 bits (68), Expect = 0.31
Identities = 18/35 (51%), Positives = 19/35 (53%), Gaps = 3/35 (8%)
Frame = -1
Query: 68 RGRGRGRGRD---VYARGRGDRGRGGRGGRGDGRG 99
R RG GRG+ RGR RGRG G GDG G
Sbjct: 287 RERGGGRGQQRGKSRRRGRRSRGRGRGGKGGDGAG 183
>TC16032 similar to BAC87886 (BAC87886) PGL-3, partial (4%)
Length = 631
Score = 39.3 bits (90), Expect = 9e-04
Identities = 32/86 (37%), Positives = 40/86 (46%), Gaps = 21/86 (24%)
Frame = +3
Query: 37 VRQTPADAESDNVPRRQPMNRFVRDDGDG-----SGRGRGRGRG------RDVYARGRGD 85
+ Q DA+ + PRR N+ G G +GRGRG RG R+ YA G+
Sbjct: 147 LNQPDVDAKDQHYPRRGYQNQRGGRGGGGRRGYSNGRGRGGSRGGYYPNGRNQYADQPGN 326
Query: 86 ---------RGRGGRGGRG-DGRGGF 101
RGRGGRG G +G GGF
Sbjct: 327 YYPRNNYNNRGRGGRGSYGNNGSGGF 404
Score = 28.1 bits (61), Expect = 2.0
Identities = 17/41 (41%), Positives = 20/41 (48%), Gaps = 6/41 (14%)
Frame = +3
Query: 79 YARGRGDRGRGGR------GGRGDGRGGFKRYGDDRTSHQD 113
Y RG RG GGR GRG RGG+ Y + R + D
Sbjct: 198 YQNQRGGRGGGGRRGYSNGRGRGGSRGGY--YPNGRNQYAD 314
>TC8052 similar to GB|AAP13431.1|30023796|BT006323 At5g02530 {Arabidopsis
thaliana;}, partial (55%)
Length = 1389
Score = 37.7 bits (86), Expect = 0.002
Identities = 23/49 (46%), Positives = 30/49 (60%)
Frame = +2
Query: 66 SGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDI 114
SG+GRG GR + G G RGRG R GRG GRG + G ++ S +D+
Sbjct: 704 SGQGRGGAFGR-LRGGGDGGRGRGHRRGRGRGRGHGRGRG-EKVSAEDL 844
Score = 33.1 bits (74), Expect = 0.061
Identities = 25/58 (43%), Positives = 26/58 (44%)
Frame = +2
Query: 42 ADAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG 99
A S+ VPR G G G GR RG RGRG R RG GRG GRG
Sbjct: 674 AFGNSNGVPR----------SGQGRGGAFGRLRGGGDGGRGRGHR-RGRGRGRGHGRG 814
Score = 27.3 bits (59), Expect = 3.4
Identities = 12/18 (66%), Positives = 12/18 (66%)
Frame = +2
Query: 63 GDGSGRGRGRGRGRDVYA 80
G G GRG GRGRG V A
Sbjct: 782 GRGRGRGHGRGRGEKVSA 835
>TC18628 weakly similar to PIR|T09527|T09527 glycine-rich protein 1 -
chickpea {Cicer arietinum;} , partial (32%)
Length = 424
Score = 37.7 bits (86), Expect = 0.002
Identities = 16/24 (66%), Positives = 18/24 (74%)
Frame = +3
Query: 82 GRGDRGRGGRGGRGDGRGGFKRYG 105
GRG GRGG GGRG G GG+ R+G
Sbjct: 15 GRGHGGRGGGGGRGGGGGGYCRHG 86
>TC9308 weakly similar to UP|Q9SQ55 (Q9SQ55) Nuclear RNA binding protein A,
partial (9%)
Length = 562
Score = 37.4 bits (85), Expect = 0.003
Identities = 27/62 (43%), Positives = 28/62 (44%), Gaps = 12/62 (19%)
Frame = +2
Query: 50 PRRQPMNRFVRDDGDGSGRGRG-RGRGRDVYA-----------RGRGDRGRGGRGGRGDG 97
PR R R D G+GRG R RG A RGRG R RG RG RG G
Sbjct: 260 PRS*SCRRRGRGDRARGGKGRGGRARGEGAPAEGAPAEGARGGRGRGGRARGPRGRRGAG 439
Query: 98 RG 99
RG
Sbjct: 440 RG 445
>TC17252 similar to UP|O04237 (O04237) Transcription factor, partial (21%)
Length = 492
Score = 37.0 bits (84), Expect = 0.004
Identities = 24/58 (41%), Positives = 29/58 (49%)
Frame = +1
Query: 41 PADAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGR 98
PA S +P Q + R +++ GRG GRG GR GRGGRGG G GR
Sbjct: 340 PAQLPSKPLPPTQAV-RESKNEASRGGRGGGRGGGRGF--------GRGGRGGSGFGR 486
Score = 29.3 bits (64), Expect = 0.89
Identities = 13/18 (72%), Positives = 13/18 (72%)
Frame = +1
Query: 83 RGDRGRGGRGGRGDGRGG 100
RG RG G GGRG GRGG
Sbjct: 409 RGGRGGGRGGGRGFGRGG 462
Score = 28.1 bits (61), Expect = 2.0
Identities = 11/23 (47%), Positives = 14/23 (60%)
Frame = +1
Query: 83 RGDRGRGGRGGRGDGRGGFKRYG 105
R + RGGRG GRGG + +G
Sbjct: 385 RESKNEASRGGRGGGRGGGRGFG 453
>TC18764 similar to UP|Q8LBJ5 (Q8LBJ5) Nucleic acid binding protein-like,
partial (35%)
Length = 848
Score = 37.0 bits (84), Expect = 0.004
Identities = 17/26 (65%), Positives = 19/26 (72%)
Frame = +3
Query: 58 FVRDDGDGSGRGRGRGRGRDVYARGR 83
F R D +G GRGRGRGRGR + RGR
Sbjct: 153 FKRVDIEGRGRGRGRGRGRGRFRRGR 230
>AV428285
Length = 239
Score = 37.0 bits (84), Expect = 0.004
Identities = 20/42 (47%), Positives = 22/42 (51%)
Frame = +3
Query: 45 ESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDR 86
+SD +R M G GRGRGRGRGR RGRG R
Sbjct: 42 DSDEEAKRSKMLELGHTSSTGKGRGRGRGRGR---GRGRGAR 158
Score = 28.1 bits (61), Expect = 2.0
Identities = 19/53 (35%), Positives = 25/53 (46%), Gaps = 1/53 (1%)
Frame = +3
Query: 61 DDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTS-HQ 112
DDG+ S R + ++ +GRG GRG GRG R + TS HQ
Sbjct: 30 DDGNDSDEEAKRSKMLELGHTSSTGKGRGRGRGRGRGRGRGARTVEKETSVHQ 188
>TC18335 similar to UP|Q7XJ13 (Q7XJ13) Chloroplast nucleoid DNA-binding
protein-like protein, partial (19%)
Length = 588
Score = 36.6 bits (83), Expect = 0.006
Identities = 22/57 (38%), Positives = 28/57 (48%), Gaps = 6/57 (10%)
Frame = +3
Query: 53 QPMNRFVRDDGDGS------GRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKR 103
+P+N + +DG+ S GRGRGRGRG Y G + G G G GG R
Sbjct: 405 KPLNEY-EEDGEXSPXMPXXGRGRGRGRGXXFYNNGGMEYGGGDGWDXGXXYGGXSR 572
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,175,582
Number of Sequences: 28460
Number of extensions: 49735
Number of successful extensions: 1137
Number of sequences better than 10.0: 204
Number of HSP's better than 10.0 without gapping: 651
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 901
length of query: 310
length of database: 4,897,600
effective HSP length: 90
effective length of query: 220
effective length of database: 2,336,200
effective search space: 513964000
effective search space used: 513964000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC147483.4


