

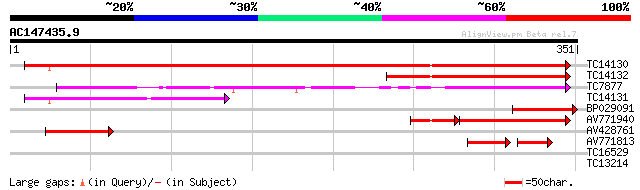
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147435.9 + phase: 0
(351 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14130 similar to UP|ATPG_PEA (P28552) ATP synthase gamma chain... 452 e-128
TC14132 homologue to UP|ATP1_ARATH (Q01908) ATP synthase gamma c... 161 2e-40
TC7877 similar to UP|ATP3_IPOBA (P26360) ATP synthase gamma chai... 111 2e-25
TC14131 similar to UP|ATPG_PEA (P28552) ATP synthase gamma chain... 104 3e-23
BP029091 73 6e-14
AV771940 56 2e-12
AV428761 57 4e-09
AV771813 42 2e-07
TC16529 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, p... 27 5.1
TC13214 similar to GB|AAP31965.1|30387599|BT006621 At4g21540 {Ar... 27 6.7
>TC14130 similar to UP|ATPG_PEA (P28552) ATP synthase gamma chain,
chloroplast precursor , partial (94%)
Length = 1527
Score = 452 bits (1162), Expect = e-128
Identities = 232/341 (68%), Positives = 276/341 (80%), Gaps = 3/341 (0%)
Frame = +3
Query: 10 RIPSKPHFPQHFQI---RCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSR 66
++PS+ P + +CG+RE+R RI SVK TQKITEAMKLVAAA+VRRAQEAV+N R
Sbjct: 195 QLPSQNSPPSRISVSPVQCGLRELRTRIESVKNTQKITEAMKLVAAAKVRRAQEAVVNGR 374
Query: 67 PFSEAFAETLHSINQSLQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEA 126
PFSE E L+SIN+ LQ +DV VPLT +RPVK VAL+V TGDRGLCGGFNN + KKAEA
Sbjct: 375 PFSETLVEVLYSINEQLQTEDVDVPLTKIRPVKKVALVVCTGDRGLCGGFNNFILKKAEA 554
Query: 127 RVMELKNLGINCVVISVGKKGSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFV 186
R+ ELK LG+ +ISVGKKG+SYF R ++ VDRF++ PT K+AQ IADDVFSLFV
Sbjct: 555 RIKELKELGLEYTIISVGKKGNSYFIRRPYIPVDRFLEGGTLPTAKEAQAIADDVFSLFV 734
Query: 187 TEEVDKVELVYTKFVSLVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGK 246
+EEVDKVEL+YTKFVSLV+ +PVI TLLPLS KGE+ D+NGN VD EDE FRLT+K+GK
Sbjct: 735 SEEVDKVELLYTKFVSLVKSDPVIHTLLPLSPKGEICDINGNCVDAAEDELFRLTTKEGK 914
Query: 247 LALKRDVKKKKMKDGFVPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGA 306
L ++RD + K D F P+++FEQDP QILDA++PLYLNSQVL+ALQESLASELAARM A
Sbjct: 915 LTVERDTIRTKTAD-FSPILQFEQDPVQILDALLPLYLNSQVLRALQESLASELAARMSA 1091
Query: 307 MSNATDNAVELTKELSVAYNRERQAKITGEILEIVAGAEAL 347
MSNA+DNA +L K LS YNR+RQAKITGEILEIVAGA AL
Sbjct: 1092MSNASDNASDLKKSLSTVYNRQRQAKITGEILEIVAGANAL 1214
>TC14132 homologue to UP|ATP1_ARATH (Q01908) ATP synthase gamma chain 1,
chloroplast precursor , partial (30%)
Length = 541
Score = 161 bits (407), Expect = 2e-40
Identities = 83/114 (72%), Positives = 97/114 (84%)
Frame = +1
Query: 234 EDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVMEFEQDPAQILDAMMPLYLNSQVLKALQ 293
EDE FRLT+K+GKL ++RD + K D F P+++FEQDP QILDA++PLYLNSQVL+ALQ
Sbjct: 1 EDELFRLTTKEGKLTVERDTFRTKTAD-FSPILQFEQDPVQILDALLPLYLNSQVLRALQ 177
Query: 294 ESLASELAARMGAMSNATDNAVELTKELSVAYNRERQAKITGEILEIVAGAEAL 347
ESLASELAARM AMSNA+DNA +L K LS YNR+RQAKITGEILEIVAGA AL
Sbjct: 178 ESLASELAARMSAMSNASDNASDLKKSLSTVYNRQRQAKITGEILEIVAGANAL 339
>TC7877 similar to UP|ATP3_IPOBA (P26360) ATP synthase gamma chain,
mitochondrial precursor , partial (91%)
Length = 1420
Score = 111 bits (277), Expect = 2e-25
Identities = 91/322 (28%), Positives = 155/322 (47%), Gaps = 4/322 (1%)
Frame = +2
Query: 30 IRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQSLQNDDVV 89
+R+R+ SVK QKIT+AMK+VAA+++R Q NSR + F L
Sbjct: 281 VRNRMKSVKNIQKITKAMKMVAASKLRAVQTRAENSRGLWQPFTALLGDA---------- 430
Query: 90 VPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGIN--CVVISVGKKG 147
T+V VK ++ ++ D+GLCGG N++ K + R + N G + + +G+K
Sbjct: 431 ---TSV-DVKKNVIVTVSSDKGLCGGINSTSVKIS--RALSKLNAGPDKETKYVILGEKA 592
Query: 148 SSYFNRSGFVEVDRFIDNVGFPTTKDAQI--IADDVFSLFVTEEVDKVELVYTKFVSLVR 205
+ R ++ + + Q+ +ADD+ E D + +V+ KF S+V+
Sbjct: 593 KAQLIRDSKQDILLSLTELQKNPLNYTQVSVLADDILK---NVEFDALRIVFNKFHSVVQ 763
Query: 206 FNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPV 265
F P + T+L S +++E E T G L ++ +++ G
Sbjct: 764 FYPTVSTVL--------------SPEIVERE----TESGGMLG---ELDSYEIEGG---- 868
Query: 266 MEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELSVAY 325
++IL + + + A+ E+ SE ARM AM +++ NA ++ L++ Y
Sbjct: 869 ----DSKSEILQNLSEFQFSCVMFNAVLENACSEQGARMSAMDSSSRNAGDMLDRLTLTY 1036
Query: 326 NRERQAKITGEILEIVAGAEAL 347
NR RQA IT E++EI++GA AL
Sbjct: 1037NRTRQASITTELIEIISGASAL 1102
>TC14131 similar to UP|ATPG_PEA (P28552) ATP synthase gamma chain,
chloroplast precursor , partial (40%)
Length = 655
Score = 104 bits (259), Expect = 3e-23
Identities = 60/131 (45%), Positives = 79/131 (59%), Gaps = 4/131 (3%)
Frame = +3
Query: 10 RIPSKPHFPQHFQI---RCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSR 66
++PS+ P + +CG+R + RI SVK TQKITE MKLV AA+VR+ QEAV+N R
Sbjct: 162 QLPSQNSPPSRISVSPVQCGLRXLXTRIESVKNTQKITEXMKLVGAAKVRKTQEAVVNGR 341
Query: 67 PFSEAFAE-TLHSINQSLQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAE 125
P + + + S N S + PLT +K L+V TG RGLCGGFNN + KKAE
Sbjct: 342 PSXKTSLKFSTASTNSSKLK--ISSPLTRSELLKKGGLVVCTGXRGLCGGFNNFILKKAE 515
Query: 126 ARVMELKNLGI 136
+ EL LG+
Sbjct: 516 GXIKELXELGL 548
>BP029091
Length = 447
Score = 73.2 bits (178), Expect = 6e-14
Identities = 36/40 (90%), Positives = 38/40 (95%)
Frame = -3
Query: 312 DNAVELTKELSVAYNRERQAKITGEILEIVAGAEALRPID 351
DNA+ELTK LSVAYNRERQAKITGE+LEIVAGAEAL PID
Sbjct: 445 DNAIELTKNLSVAYNRERQAKITGELLEIVAGAEALTPID 326
>AV771940
Length = 512
Score = 55.8 bits (133), Expect(2) = 2e-12
Identities = 39/71 (54%), Positives = 46/71 (63%), Gaps = 2/71 (2%)
Frame = -1
Query: 279 MMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKEL-SVAYNRERQAKITG-E 336
++PLYLNSQVL+ALQESLASELAARM A L +EL + Y AKITG +
Sbjct: 419 LLPLYLNSQVLRALQESLASELAARMSADE*CE**C**LEEELVNSVYTARVTAKITGPQ 240
Query: 337 ILEIVAGAEAL 347
IV+GA AL
Sbjct: 239 FWNIVSGANAL 207
Score = 32.3 bits (72), Expect(2) = 2e-12
Identities = 15/30 (50%), Positives = 21/30 (70%)
Frame = -3
Query: 249 LKRDVKKKKMKDGFVPVMEFEQDPAQILDA 278
++RD + D F P+++FEQDP QILDA
Sbjct: 507 VERDTIRTDTAD-FSPILQFEQDPVQILDA 421
>AV428761
Length = 293
Score = 57.4 bits (137), Expect = 4e-09
Identities = 26/42 (61%), Positives = 35/42 (82%)
Frame = +3
Query: 23 IRCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVIN 64
++CG+ E+R +I S K TQK T+A+KLV AA+VRRAQEAV+N
Sbjct: 165 VQCGLIELRTQIESTKNTQKTTDAIKLVVAAKVRRAQEAVVN 290
>AV771813
Length = 496
Score = 42.0 bits (97), Expect(2) = 2e-07
Identities = 22/27 (81%), Positives = 23/27 (84%)
Frame = -3
Query: 284 LNSQVLKALQESLASELAARMGAMSNA 310
LNS VL+AL ESL SELAARM AMSNA
Sbjct: 494 LNSHVLRALPESLPSELAARMSAMSNA 414
Score = 28.9 bits (63), Expect(2) = 2e-07
Identities = 14/22 (63%), Positives = 15/22 (67%)
Frame = -2
Query: 315 VELTKELSVAYNRERQAKITGE 336
V L K LS YNR+RQAKI E
Sbjct: 399 VTLKKSLSTVYNRQRQAKIHQE 334
>TC16529 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(22%)
Length = 761
Score = 26.9 bits (58), Expect = 5.1
Identities = 15/40 (37%), Positives = 19/40 (47%), Gaps = 10/40 (25%)
Frame = -3
Query: 3 STTITNGRIPSKPHFP----------QHFQIRCGIREIRD 32
STT+T+ RIP H P QH Q C IR + +
Sbjct: 594 STTLTDTRIPMGIHDPNRPVVQWSEIQHIQCPCSIRRVAE 475
>TC13214 similar to GB|AAP31965.1|30387599|BT006621 At4g21540 {Arabidopsis
thaliana;}, partial (7%)
Length = 484
Score = 26.6 bits (57), Expect = 6.7
Identities = 16/55 (29%), Positives = 27/55 (49%), Gaps = 5/55 (9%)
Frame = -2
Query: 37 VKTTQKITEAMKLVAAARVR-----RAQEAVINSRPFSEAFAETLHSINQSLQND 86
++ T++I E KLV RVR + V N P+S A + +++ L+ D
Sbjct: 390 LRATERIDEFTKLVTPERVRLIGGWLKNDVVTNESPWSPGGAAPISCLDEGLELD 226
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.135 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,093,346
Number of Sequences: 28460
Number of extensions: 41740
Number of successful extensions: 216
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 211
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 211
length of query: 351
length of database: 4,897,600
effective HSP length: 91
effective length of query: 260
effective length of database: 2,307,740
effective search space: 600012400
effective search space used: 600012400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC147435.9


