

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147431.3 + phase: 0 /pseudo
(659 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
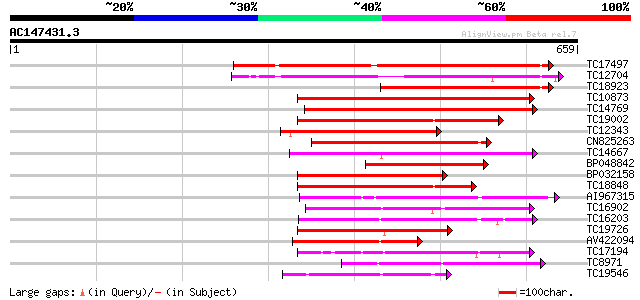
Score E
Sequences producing significant alignments: (bits) Value
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 332 2e-91
TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3,... 303 4e-83
TC18923 UP|CAE45594 (CAE45594) S-receptor kinase-like protein 1,... 242 1e-64
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 226 1e-59
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 212 2e-55
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 201 4e-52
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 201 4e-52
CN825263 199 2e-51
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 198 2e-51
BP048842 196 8e-51
BP032158 189 1e-48
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 186 8e-48
AI967315 185 2e-47
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 182 1e-46
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 180 7e-46
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 169 1e-42
AV422094 164 3e-41
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 162 2e-40
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 162 2e-40
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 154 4e-38
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 332 bits (850), Expect = 2e-91
Identities = 181/373 (48%), Positives = 239/373 (63%), Gaps = 1/373 (0%)
Frame = +1
Query: 261 KQEGTSKAKTLIIIFVSITVAVALLSCWVYSYWRKNRLSKGGMLSRTITPISFRNQVQRQ 320
K T K +++ ++ + + +L + + ++ + +G I + ++
Sbjct: 1438 KSINTKKLAGSLVVIIAFVIFITILGLAISTCIQRKKNKRGDEGEIGI----INHWKDKR 1605
Query: 321 DSFNGELPTI-PLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQG 379
+ +L TI + I +T+ FS S KLGEGGFGPVYKG L +G+E+AVKRLS TS QG
Sbjct: 1606 GDEDIDLATIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQG 1785
Query: 380 SEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWK 439
EEFKNE+ IA+LQHRNL KL G + DE + M L + + K +DW
Sbjct: 1786 MEEFKNEIKLIARLQHRNLVKLFGCSVHQDENSHANKKMKI------LLDSTRSKLVDWN 1947
Query: 440 LRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQT 499
RL II+GIARGLLYLH+DSRLR+IHRDLK SN+LLDDEMNPKISDFGLAR F DQ +
Sbjct: 1948 KRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEA 2127
Query: 500 KTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYT 559
+TKRV GTYGYM PEYA+ G FS+KSDVFSFGV+VLEII GK+ G F+ H +LL +
Sbjct: 2128 RTKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHA 2307
Query: 560 WKLWCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRMLGSDTVD 619
W+LW E + LEL+D I +E+L+ IH+ LLCVQ +RP M ++V ML + +
Sbjct: 2308 WRLWIEERPLELVDELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIVLMLNGEK-E 2484
Query: 620 LPKPTQPAFSVGR 632
LPKP PAF G+
Sbjct: 2485 LPKPRLPAFYTGK 2523
>TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3, complete
Length = 2481
Score = 303 bits (777), Expect = 4e-83
Identities = 189/419 (45%), Positives = 243/419 (57%), Gaps = 33/419 (7%)
Frame = +1
Query: 258 NPGKQEGTSKAKTLIIIFVSITVAVALLSCWVYSYWRKNRLSKGGMLSRTITPISFRNQV 317
N K G+ II ++I + +A ++C + R +GG+ +R I +
Sbjct: 1222 NIKKLAGSLAGSIAFIICITI-LGLATVTC--IRRKKNEREDEGGIETRIIN-----HWK 1377
Query: 318 QRQDSFNGELPTI-PLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETS 376
++ + +L TI + I +T+ FSES KLGEGGFGPVYKG L +G+E+AVKRLS TS
Sbjct: 1378 DKRGDEDIDLATIFDFSTISSTTNHFSESNKLGEGGFGPVYKGVLANGQEIAVKRLSNTS 1557
Query: 377 SQGSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHL 436
QG EEFKNEV IA+LQHRNL KLLG I DE +L+YE+M N SLD+ +F
Sbjct: 1558 GQGMEEFKNEVKLIARLQHRNLVKLLGCSIHHDEMLLIYEFMHNRSLDYFIF-------- 1713
Query: 437 DWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQ 496
DSRLR+IHRDLK SN+LLD EMNPKISDFGLAR F DQ
Sbjct: 1714 ---------------------DSRLRIIHRDLKTSNILLDSEMNPKISDFGLARIFTGDQ 1830
Query: 497 CQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLL 556
+ KTKRV GTYGYM+PEYA+ G FSVKSDVFSFGV+VLEII GK+ G F H ++LL
Sbjct: 1831 VEAKTKRVMGTYGYMSPEYAVHGSFSVKSDVFSFGVIVLEIISGKKIGRFCDPHHHRNLL 2010
Query: 557 LYT------------------------WKLWCEGKCLELIDPFHQKTYIESEVLKCIHIG 592
++ W+LW E + LEL+D I +E+L+ IHI
Sbjct: 2011 SHSSNFAVFLIKALRICMFENVKNRKAWRLWIEERPLELVDELLDGLAIPTEILRYIHIA 2190
Query: 593 LLCVQEDAADRPTMSTVVRMLGSDTVDLPKPTQPAFSVGR--------KSKNEDQISKN 643
LLCVQ+ RP M +VV ML + +LPKP+ PAF G SKN +++ K+
Sbjct: 2191 LLCVQQRPEYRPDMLSVVLMLNGEK-ELPKPSLPAFYTGNDDLLWPESTSKNCERVIKH 2364
>TC18923 UP|CAE45594 (CAE45594) S-receptor kinase-like protein 1, complete
Length = 2061
Score = 242 bits (618), Expect = 1e-64
Identities = 120/201 (59%), Positives = 149/201 (73%)
Frame = +1
Query: 432 KHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLART 491
+ K LDW RL II+GIARGLLYLH+DSRLR+IHRDLK SN+LLD+EMNPKISDFGLAR
Sbjct: 1387 RSKLLDWNKRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDNEMNPKISDFGLARI 1566
Query: 492 FDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEH 551
F DQ + +TKRV GTYGYM PEYA+ G FS+KSDVFSFGV+VLEII GK+ F+ H
Sbjct: 1567 FIGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIRKFYDPHH 1746
Query: 552 MQSLLLYTWKLWCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVR 611
+LL + W+LW EG LEL+D + + I +E+L+ IH+ LLCVQ RP M ++V
Sbjct: 1747 HLNLLSHAWRLWIEGSPLELVDKLFEDSIIPTEILRYIHVALLCVQRRPETRPDMLSIVL 1926
Query: 612 MLGSDTVDLPKPTQPAFSVGR 632
ML + +LPKP+ PAF G+
Sbjct: 1927 MLNGEK-ELPKPSLPAFYTGK 1986
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 226 bits (575), Expect = 1e-59
Identities = 120/278 (43%), Positives = 175/278 (62%), Gaps = 2/278 (0%)
Frame = +1
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQ 394
+ +T +F E+ +GEGGFG VYKG L G VAVK+LS QG +EF EV+ ++ L
Sbjct: 430 LADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLLH 609
Query: 395 HRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKH-LDWKLRLSIINGIARGLL 453
H NL +L+GYC +GD+++LVYEYMP SL+ HLF K L+W R+ + G ARGL
Sbjct: 610 HTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLE 789
Query: 454 YLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAP 513
YLH + VI+RDLK++N+LLD+E NPK+SDFGLA+ + RV GTYGY AP
Sbjct: 790 YLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAP 969
Query: 514 EYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCL-ELI 572
EYAM+G ++KSD++SFGV++LE++ G+R D Q+L+ + + + + ++
Sbjct: 970 EYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMV 1149
Query: 573 DPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVV 610
DP Q + + + I I +C+QE RP ++ +V
Sbjct: 1150DPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIV 1263
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 212 bits (539), Expect = 2e-55
Identities = 121/274 (44%), Positives = 165/274 (60%), Gaps = 3/274 (1%)
Frame = +3
Query: 343 SESYK--LGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLAK 400
+E YK +GEGGFG VY+GTL DG+EVAVK S TS+QG+ EF NE+ ++ +QH NL
Sbjct: 2160 TERYKTLIGEGGFGSVYRGTLNDGQEVAVKVRSSTSTQGTREFDNELNLLSAIQHENLVP 2339
Query: 401 LLGYCIEGDEKILVYEYMPNSSLDFHLFNEE-KHKHLDWKLRLSIINGIARGLLYLHEDS 459
LLGYC E D++ILVY +M N SL L+ E K K LDW RLSI G ARGL YLH
Sbjct: 2340 LLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALGAARGLAYLHTFP 2519
Query: 460 RLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYAMAG 519
VIHRD+K+SN+LLD M K++DFG ++ ++ + V GT GY+ PEY
Sbjct: 2520 GRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKTQ 2699
Query: 520 LFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLELIDPFHQKT 579
S KSDVFSFGV++LEI+ G+ + SL+ + K E++DP +
Sbjct: 2700 QLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEWATPYIRGSKVDEIVDPGIKGG 2879
Query: 580 YIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML 613
Y + + + + L C++ + RP+M +VR L
Sbjct: 2880 YHAEAMWRVVEVALQCLEPFSTYRPSMVAIVREL 2981
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 201 bits (510), Expect = 4e-52
Identities = 108/241 (44%), Positives = 150/241 (61%), Gaps = 1/241 (0%)
Frame = +1
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQ 394
+ +T++F+ KLGEGGFG VY G L DG ++AVKRL S++ EF EV +A+++
Sbjct: 43 LHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEILARVR 222
Query: 395 HRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHK-HLDWKLRLSIINGIARGLL 453
H+NL L GYC EG E+++VY+YMPN SL HL + + LDW R++I G A G++
Sbjct: 223 HKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGSAEGIV 402
Query: 454 YLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAP 513
YLH + +IHRD+KASNVLLD + +++DFG A+ D T RV GT GY+AP
Sbjct: 403 YLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLI-PDGATHVTTRVKGTLGYLAP 579
Query: 514 EYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLELID 573
EYAM G + DVFSFG+L+LE+ GK+ + S +S+ + L C K E D
Sbjct: 580 EYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALPLACAKKFTEFAD 759
Query: 574 P 574
P
Sbjct: 760 P 762
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 201 bits (510), Expect = 4e-52
Identities = 99/191 (51%), Positives = 128/191 (66%), Gaps = 4/191 (2%)
Frame = +3
Query: 315 NQVQRQDSFNG----ELPTIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVK 370
++ Q +D E T + +T +F KLGEGGFGPV+KG L DGRE+AVK
Sbjct: 150 SEAQNEDDIQNIAAKEHKTFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLNDGREIAVK 329
Query: 371 RLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNE 430
+LS S+QG +F NE + ++QHRN+ L GYC G EK+LVYEY+P SLD LF
Sbjct: 330 KLSRRSNQGRTQFINEAKLLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLLFRS 509
Query: 431 EKHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLAR 490
+K + LDWK R II+G+ARGLLYLHEDS +IHRD+KA+N+LLD++ PKI+DFGLAR
Sbjct: 510 QKKEQLDWKRRFDIISGVARGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADFGLAR 689
Query: 491 TFDKDQCQTKT 501
F +DQ T
Sbjct: 690 IFPEDQTHVNT 722
>CN825263
Length = 663
Score = 199 bits (505), Expect = 2e-51
Identities = 106/210 (50%), Positives = 137/210 (64%), Gaps = 1/210 (0%)
Frame = +1
Query: 352 GFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEK 411
GFG VYKG L DGR+VAVK L +G EF EV +++L HRNL KL+G CIE +
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTR 180
Query: 412 ILVYEYMPNSSLDFHLFNEEKHKH-LDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKA 470
L+YE +PN S++ HL +K LDW R+ I G ARGL YLHEDS VIHRD K+
Sbjct: 181 CLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFKS 360
Query: 471 SNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSF 530
SN+LL+ + PK+SDFGLART + + + V GT+GY+APEYAM G VKSDV+S+
Sbjct: 361 SNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYSY 540
Query: 531 GVLVLEIIYGKRNGDFFLSEHMQSLLLYTW 560
GV++LE++ G + D LS+ L TW
Sbjct: 541 GVVLLELLTGTKPVD--LSQPPGQENLVTW 624
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 198 bits (504), Expect = 2e-51
Identities = 112/295 (37%), Positives = 168/295 (55%), Gaps = 7/295 (2%)
Frame = +2
Query: 326 ELPTIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSS-QGSEEFK 384
E+P + L +++ TD+F +GEG +G VY TL DG VAVK+L +S + + EF
Sbjct: 311 EVPALSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNEFL 490
Query: 385 NEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNE------EKHKHLDW 438
+V +++L++ N +L GYC+EG+ ++L YE+ SL L + LDW
Sbjct: 491 TQVSMVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDW 670
Query: 439 KLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQ 498
R+ I ARGL YLHE + +IHRD+++SNVL+ ++ KI+DF L+ +
Sbjct: 671 IQRVRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAAR 850
Query: 499 TKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLY 558
+ RV GT+GY APEYAM G + KSDV+SFGV++LE++ G++ D + QSL+ +
Sbjct: 851 LHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTW 1030
Query: 559 TWKLWCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML 613
E K + +DP + Y V K + LCVQ +A RP MS VV+ L
Sbjct: 1031ATPRLSEDKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKAL 1195
>BP048842
Length = 524
Score = 196 bits (499), Expect = 8e-51
Identities = 95/143 (66%), Positives = 115/143 (79%)
Frame = -3
Query: 414 VYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNV 473
+YE+M N SL++ +F+ + K +DW RL II+GIARGLLYLH+DSRLR+IHRDLK SN+
Sbjct: 522 IYEFMHNRSLNYFIFDSTRSKLVDWNKRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNI 343
Query: 474 LLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVL 533
LLDDEMNPKISDFGLAR F DQ + +TKRV GTYGYM PEYA+ G FS+KSDVFSFGV+
Sbjct: 342 LLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVI 163
Query: 534 VLEIIYGKRNGDFFLSEHMQSLL 556
VLEII GK+ G F+ H +LL
Sbjct: 162 VLEIISGKKIGRFYDPHHHLNLL 94
>BP032158
Length = 555
Score = 189 bits (480), Expect = 1e-48
Identities = 95/176 (53%), Positives = 130/176 (72%), Gaps = 1/176 (0%)
Frame = +2
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQ 394
I+ +T++F + K+GEGGFGPVYKG L +G +AVK+LS S QG+ EF NE+ I+ LQ
Sbjct: 29 IKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSSKSKQGNREFINEIGMISALQ 208
Query: 395 HRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLF-NEEKHKHLDWKLRLSIINGIARGLL 453
H NL KL G CIEG++ +LVYEYM N+SL LF NEE+ +L+W+ R+ I GIA+GL
Sbjct: 209 HPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKICVGIAKGLA 388
Query: 454 YLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYG 509
YLHE+SRL+++HRD+KA+NVLLD ++ KISDFGLA+ +++ T R+ GT G
Sbjct: 389 YLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHIST-RIAGTIG 553
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 186 bits (473), Expect = 8e-48
Identities = 94/209 (44%), Positives = 135/209 (63%), Gaps = 1/209 (0%)
Frame = +2
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQ 394
+ +T FS+ KLGEGGFG VY G DG ++AVK+L +S+ EF EV + +++
Sbjct: 281 LHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEVLGRVR 460
Query: 395 HRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNE-EKHKHLDWKLRLSIINGIARGLL 453
H+NL L GYC+ D++++VY+YMPN SL HL + L+W+ R+ I G A G+L
Sbjct: 461 HKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIGSAEGIL 640
Query: 454 YLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAP 513
YLH + +IHRD+KASNVLL+ + P ++DFG A+ + T RV GT GY+AP
Sbjct: 641 YLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLI-PEGVSHMTTRVKGTLGYLAP 817
Query: 514 EYAMAGLFSVKSDVFSFGVLVLEIIYGKR 542
EYAM G S DV+SFG+L+LE++ G++
Sbjct: 818 EYAMWGKVSESCDVYSFGILLLELVTGRK 904
>AI967315
Length = 1308
Score = 185 bits (469), Expect = 2e-47
Identities = 113/304 (37%), Positives = 170/304 (55%), Gaps = 2/304 (0%)
Frame = +1
Query: 338 STDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSET--SSQGSEEFKNEVIFIAKLQH 395
+T+ FS +G+GG+ VYKG L G E+AVKRL+ T + +EF E+ I + H
Sbjct: 136 ATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEIGTIGHVCH 315
Query: 396 RNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGLLYL 455
N+ LLG CI+ LV+E S+ L ++EK LDWK R I+ G ARGL YL
Sbjct: 316 SNVMPLLGCCIDNG-LYLVFELSTVGSVA-SLIHDEKMAPLDWKTRYKIVLGTARGLHYL 489
Query: 456 HEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEY 515
H+ + R+IHRD+KASN+LL ++ P+ISDFGLA+ + GT+G++APEY
Sbjct: 490 HKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIEGTFGHLAPEY 669
Query: 516 AMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLELIDPF 575
M G+ K+DVF+FGV +LE+I G++ D H QSL + + + + +L+DP
Sbjct: 670 YMHGVVDEKTDVFAFGVFLLEVISGRKPVD---GSH-QSLHTWAKPILSKWEIEKLVDPR 837
Query: 576 HQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRMLGSDTVDLPKPTQPAFSVGRKSK 635
+ Y ++ + LC++ + RPTMS V+ ++ +D + P K +
Sbjct: 838 LEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVMEEGEMDKERWKMP------KEE 999
Query: 636 NEDQ 639
EDQ
Sbjct: 1000EEDQ 1011
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 182 bits (462), Expect = 1e-46
Identities = 103/270 (38%), Positives = 156/270 (57%), Gaps = 3/270 (1%)
Frame = +3
Query: 344 ESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLG 403
E+ LG GGFG VY G + G +VA+KR + S QG EF+ E+ ++KL+HR+L L+G
Sbjct: 3 EALLLGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIG 182
Query: 404 YCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGLLYLHEDSRLRV 463
YC E E ILVY++M +L HL+ +K L WK RL I G ARGL YLH ++ +
Sbjct: 183 YCEENTEMILVYDHMAYGTLREHLYKTQK-PPLPWKQRLEICIGAARGLHYLHTGAKYTI 359
Query: 464 IHRDLKASNVLLDDEMNPKISDFGLAR---TFDKDQCQTKTKRVFGTYGYMAPEYAMAGL 520
IHRD+K +N+LLD++ K+SDFGL++ T D T K G++GY+ PEY
Sbjct: 360 IHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVK---GSFGYLDPEYFRRQQ 530
Query: 521 FSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLELIDPFHQKTY 580
+ KSDV+SFGV++ EI+ + + L++ SL + + +G +++DP+ +
Sbjct: 531 LTDKSDVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCYNKGILDQILDPYLKGKI 710
Query: 581 IESEVLKCIHIGLLCVQEDAADRPTMSTVV 610
K + CV + +RP+M V+
Sbjct: 711 APECFKKFAETAMKCVSDQGIERPSMGDVL 800
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 180 bits (456), Expect = 7e-46
Identities = 107/288 (37%), Positives = 159/288 (55%), Gaps = 10/288 (3%)
Frame = +2
Query: 336 EQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRL-SETSSQGSEEFKNEVIFIAKLQ 394
E + E +G+GG G VY+G++P+G +VA+KRL + S + F+ E+ + K++
Sbjct: 2174 EDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIR 2353
Query: 395 HRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGLLY 454
HRN+ +LLGY D +L+YEYMPN SL L K HL W++R I ARGL Y
Sbjct: 2354 HRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGA-KGGHLRWEMRYKIAVEAARGLCY 2530
Query: 455 LHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPE 514
+H D +IHRD+K++N+LLD + ++DFGLA+ + G+YGY+APE
Sbjct: 2531 MHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPE 2710
Query: 515 YAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTW--KLWCE------- 565
YA KSDV+SFGV++LE+I G++ + E + + W K E
Sbjct: 2711 YAYTLKVDEKSDVYSFGVVLLELIIGRKP----VGEFGDGVDIVGWVNKTMSELSQPSDT 2878
Query: 566 GKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML 613
L ++DP Y + V+ +I ++CV+E RPTM VV ML
Sbjct: 2879 ALVLAVVDP-RLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 3019
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 169 bits (429), Expect = 1e-42
Identities = 85/185 (45%), Positives = 126/185 (67%), Gaps = 5/185 (2%)
Frame = +3
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLPDGR-EVAVKRLSETSSQGSEEFKNEVIFIAKL 393
++++T++F E +KLG+GG+G VY+G LP + EVAVK S + +++F +E+I I +L
Sbjct: 147 LKKATNNFDEKHKLGQGGYGVVYRGMLPKEKLEVAVKMFSRDKMKSTDDFLSELIIINRL 326
Query: 394 QHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHK---HLDWKLRLSIINGIAR 450
+H++L +L G+C + +LVY+YMPN SLD H+F EE L W LR II+G+A
Sbjct: 327 RHKHLVRLQGWCHKNGVLLLVYDYMPNGSLDSHIFCEEGGTITTPLSWPLRYKIISGVAS 506
Query: 451 GLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQ-TKTKRVFGTYG 509
L YLH + +V+HRDLKASN++LD E N K+ DFGLAR + ++ T+ + V GT G
Sbjct: 507 ALNYLHNEYDQKVVHRDLKASNIMLDSEFNAKLGDFGLARALENEKISYTELEGVHGTMG 686
Query: 510 YMAPE 514
Y+APE
Sbjct: 687 YIAPE 701
>AV422094
Length = 462
Score = 164 bits (416), Expect = 3e-41
Identities = 83/151 (54%), Positives = 107/151 (69%)
Frame = +2
Query: 329 TIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVI 388
T + ++ +T+DF+ KLGEGGFGPVYKG L DG +AVK+LS S QG +F E+
Sbjct: 11 TFSYSELKNATNDFNIDNKLGEGGFGPVYKGILNDGTVIAVKQLSLGSHQGKSQFIAEIA 190
Query: 389 FIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGI 448
I+ +QHRNL KL G CIEG +++LVYEY+ N SLD LF K L+W R I G+
Sbjct: 191 TISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQALFG--KALSLNWSTRYDICLGV 364
Query: 449 ARGLLYLHEDSRLRVIHRDLKASNVLLDDEM 479
ARGL YLHE+SRLR++HRD+KASN+LLD E+
Sbjct: 365 ARGLTYLHEESRLRIVHRDVKASNILLDHEL 457
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 162 bits (410), Expect = 2e-40
Identities = 101/283 (35%), Positives = 160/283 (55%), Gaps = 7/283 (2%)
Frame = +1
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQ 394
+ ++T++FS K+G+GGFG VY L G++ A+K++ Q S EF E+ + +
Sbjct: 1066 LAKATNNFSLDNKIGQGGFGAVYYAELR-GKKTAIKKMDV---QASTEFLCELKVLTHVH 1233
Query: 395 HRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGLLY 454
H NL +L+GYC+EG LVYE++ N +L +L K L W R+ I ARGL Y
Sbjct: 1234 HLNLVRLIGYCVEGS-LFLVYEHIDNGNLGQYLHGSGKEP-LPWSSRVQIALDAARGLEY 1407
Query: 455 LHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPE 514
+HE + IHRD+K++N+L+D + K++DFGL + + +T R+ GT+GYM PE
Sbjct: 1408 IHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQT-RLVGTFGYMPPE 1584
Query: 515 YAMAGLFSVKSDVFSFGVLVLEIIYGK----RNGDFFLSEHMQSLLLYTWKLWCEGKC-- 568
YA G S K DV++FGV++ E+I K + G+ ++E + L+ L C
Sbjct: 1585 YAQYGDISPKIDVYAFGVVLFELISAKNAVLKTGE-LVAESKGLVALFEEALNKSDPCDA 1761
Query: 569 -LELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVV 610
+L+DP + Y VLK +G C +++ RP+M ++V
Sbjct: 1762 LRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLV 1890
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 162 bits (409), Expect = 2e-40
Identities = 93/239 (38%), Positives = 140/239 (57%), Gaps = 2/239 (0%)
Frame = +3
Query: 386 EVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSII 445
EV +AK+ HRNL KLLG+ +G+E+IL+ EY+PN +L HL + + K LD+ RL I
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHL-DGLRGKILDFNQRLEIA 182
Query: 446 NGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLART--FDKDQCQTKTKR 503
+A GL YLH + ++IHRD+K+SN+LL + M K++DFG AR + DQ TK
Sbjct: 183 IDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTK- 359
Query: 504 VFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLW 563
V GT GY+ PEY + KSDV+SFG+L+LEI+ G+R + + + L + ++ +
Sbjct: 360 VKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRKY 539
Query: 564 CEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRMLGSDTVDLPK 622
EG +EL+DP ++ ++K + + C DRP M +V L + D K
Sbjct: 540 NEGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSVGEQLWAIRADYLK 716
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 154 bits (389), Expect = 4e-38
Identities = 85/198 (42%), Positives = 120/198 (59%), Gaps = 2/198 (1%)
Frame = +1
Query: 318 QRQDSFNGE--LPTIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSET 375
Q D F +P I+++T +F+ LG+G FG VYK T+P G VAVK L+
Sbjct: 157 QNNDQFASPSGIPKYSYKDIQKATQNFTTI--LGQGSFGTVYKATMPTGEVVAVKVLAPN 330
Query: 376 SSQGSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKH 435
S QG EF+ EV + +L HRNL L+G+C++ ++ILVY++M N SL L+ EE K
Sbjct: 331 SKQGEHEFQTEVHLLGRLHHRNLVNLVGFCVDKGQRILVYQFMSNGSLANLLYGEE--KE 504
Query: 436 LDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKD 495
L W RL I I+ G+ YLHE + VIHRDLK++N+LLDD M ++DFGL++ ++
Sbjct: 505 LSWDERLQIAMDISHGIEYLHEGAVPPVIHRDLKSANILLDDSMRAMVADFGLSK---EE 675
Query: 496 QCQTKTKRVFGTYGYMAP 513
+ + GTYGYM P
Sbjct: 676 IFDGRNSGLKGTYGYMDP 729
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,330,445
Number of Sequences: 28460
Number of extensions: 189891
Number of successful extensions: 1588
Number of sequences better than 10.0: 331
Number of HSP's better than 10.0 without gapping: 1383
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1402
length of query: 659
length of database: 4,897,600
effective HSP length: 96
effective length of query: 563
effective length of database: 2,165,440
effective search space: 1219142720
effective search space used: 1219142720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC147431.3


