

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147363.5 - phase: 0
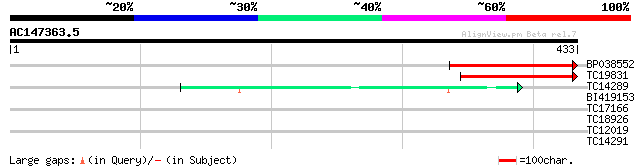
(433 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP038552 167 3e-42
TC19831 similar to UP|Q9LJ19 (Q9LJ19) ESTs C97134(C52296) (Hydr... 130 3e-31
TC14289 similar to UP|LEU1_SOYBN (Q39891) Probable 2-isopropylma... 46 1e-05
BI419153 29 1.3
TC17166 homologue to UP|Q84L15 (Q84L15) Cytosolic glutamine synt... 28 2.9
TC18926 similar to AAQ63968 (AAQ63968) VAP27-1, partial (36%) 28 2.9
TC12019 27 8.6
TC14291 similar to UP|LEU1_SOYBN (Q39891) Probable 2-isopropylma... 27 8.6
>BP038552
Length = 523
Score = 167 bits (423), Expect = 3e-42
Identities = 83/97 (85%), Positives = 91/97 (93%)
Frame = -2
Query: 337 DTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLNGLGIKTNVD 396
DTYG SLP ILVSL+MGISA+DSSV+ LGGCPYAKGASGNVATEDVVYMLNGLG+KTNVD
Sbjct: 522 DTYGPSLPYILVSLRMGISALDSSVSVLGGCPYAKGASGNVATEDVVYMLNGLGVKTNVD 343
Query: 397 IGKLMSAGDFIGKQLGRPSGSKTAIALNRVTADASKI 433
+GKL+SAGDFI K LGRPSGSKTA+AL RVT+DASKI
Sbjct: 342 LGKLLSAGDFISKHLGRPSGSKTAVALGRVTSDASKI 232
>TC19831 similar to UP|Q9LJ19 (Q9LJ19) ESTs C97134(C52296)
(Hydroxymethylglutaryl-CoA lyase) (HMG-CoA lyase) (HL)
(3-hydroxy-3-methylglutarate-CoA lyase) , partial (19%)
Length = 488
Score = 130 bits (328), Expect = 3e-31
Identities = 66/89 (74%), Positives = 76/89 (85%)
Frame = +2
Query: 345 NILVSLQMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAG 404
N LVSLQMGIS VDSSV+GLG CPYA+GA+GNVATEDVVYML GLG+KT+VD+GKLM AG
Sbjct: 5 NTLVSLQMGISTVDSSVSGLGXCPYAQGATGNVATEDVVYMLTGLGVKTHVDLGKLMLAG 184
Query: 405 DFIGKQLGRPSGSKTAIALNRVTADASKI 433
DFI K LGR SGSK A A ++VT AS++
Sbjct: 185 DFICKHLGRASGSKAATASSKVTNHASQL 271
>TC14289 similar to UP|LEU1_SOYBN (Q39891) Probable 2-isopropylmalate
synthase (Alpha-isopropylmalate synthase) (Alpha-IPM
synthetase) (Late nodulin 56) (N-56) , partial (85%)
Length = 1822
Score = 45.8 bits (107), Expect = 1e-05
Identities = 55/272 (20%), Positives = 108/272 (39%), Gaps = 11/272 (4%)
Frame = +3
Query: 131 PEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEA-------TSFVSPKWV 183
P +V+I++ RDG Q+ + K+E +LA G+ +IEA F++ K +
Sbjct: 120 PTYVRILDTTLRDGEQSPGAAMTCVQKLETARQLAKLGVDIIEAGFPCASKQDFMAVKMI 299
Query: 184 -PQLADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINC 242
++ + D V + G+ +EA A + F + S + +
Sbjct: 300 AEEVGNCVDGNGYVPVITGVSRCNEKDIATAWEALKHAKRPRLRTFIATSPIHMEYKLRK 479
Query: 243 SIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEIS 302
S ++ L R + + A+ L G G + + + G ++
Sbjct: 480 SKDQVLETARNMVKFARSL-----GCTDIQFGAEDAARSDREFLYQIFGEVIKAGATTLT 644
Query: 303 LGDTIGVGTPGTVVPMLLAVMAIVPTEKLAV---HFHDTYGQSLPNILVSLQMGISAVDS 359
+ DT+G+ P ++ + A P + A+ H H+ G + N + + G ++
Sbjct: 645 IPDTVGIAMPFEYGKLIADIKANTPGIENAIISTHCHNDLGLATSNTIEGARYGARQLEV 824
Query: 360 SVAGLGGCPYAKGASGNVATEDVVYMLNGLGI 391
++ G+G +GN + E+VV L GI
Sbjct: 825 TINGIG------ERAGNASFEEVVMALTCRGI 902
>BI419153
Length = 499
Score = 29.3 bits (64), Expect = 1.3
Identities = 14/42 (33%), Positives = 24/42 (56%)
Frame = +3
Query: 133 FVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEA 174
+V+I++ RDG Q + D K+E+ +L G+ +IEA
Sbjct: 153 YVRILDTTLRDGEQAPGAAMTRDQKLEVARQLVKLGVDIIEA 278
>TC17166 homologue to UP|Q84L15 (Q84L15) Cytosolic glutamine synthetase,
partial (49%)
Length = 597
Score = 28.1 bits (61), Expect = 2.9
Identities = 12/25 (48%), Positives = 15/25 (60%)
Frame = +1
Query: 328 TEKLAVHFHDTYGQSLPNILVSLQM 352
T +L HFH T G LP VSL++
Sbjct: 355 THQLESHFHQTIGTMLPKFSVSLRL 429
>TC18926 similar to AAQ63968 (AAQ63968) VAP27-1, partial (36%)
Length = 419
Score = 28.1 bits (61), Expect = 2.9
Identities = 13/36 (36%), Positives = 16/36 (44%)
Frame = -2
Query: 45 EGRSCSTSNSCNEDNEDYTAETYPWKRQTRDMSRGD 80
EG CS+SN N D +PW + R R D
Sbjct: 154 EGSQCSSSNRDRRRNGDCAVRRFPWPYEMR*DGRRD 47
>TC12019
Length = 510
Score = 26.6 bits (57), Expect = 8.6
Identities = 10/18 (55%), Positives = 12/18 (66%)
Frame = +3
Query: 14 PSMSTMDRVQRFSSGCCR 31
PS S +DRV F GCC+
Sbjct: 285 PSPSLLDRVTSFGLGCCK 338
>TC14291 similar to UP|LEU1_SOYBN (Q39891) Probable 2-isopropylmalate
synthase (Alpha-isopropylmalate synthase) (Alpha-IPM
synthetase) (Late nodulin 56) (N-56) , partial (40%)
Length = 902
Score = 26.6 bits (57), Expect = 8.6
Identities = 19/88 (21%), Positives = 39/88 (43%), Gaps = 6/88 (6%)
Frame = +3
Query: 331 LAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYML---- 386
++ H H+ G + N + + G ++ ++ G+G +GN + E+VV L
Sbjct: 18 ISTHCHNDLGLATANTIEGARAGARQLEVTINGIG------ERAGNASLEEVVMALACRG 179
Query: 387 NGL--GIKTNVDIGKLMSAGDFIGKQLG 412
N + G+ T V+ ++ + K G
Sbjct: 180 NHILGGLHTRVNTRHILKTSKMVEKYTG 263
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,534,559
Number of Sequences: 28460
Number of extensions: 82813
Number of successful extensions: 425
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 422
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 424
length of query: 433
length of database: 4,897,600
effective HSP length: 93
effective length of query: 340
effective length of database: 2,250,820
effective search space: 765278800
effective search space used: 765278800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC147363.5


