

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147202.9 + phase: 0 /pseudo
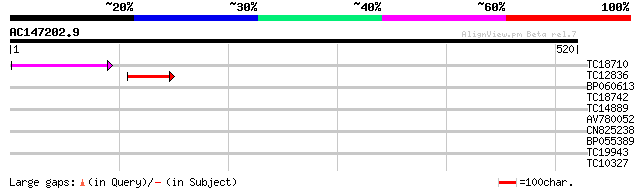
(520 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18710 83 1e-16
TC12836 GB|CAA71302.1|2292921|LJPANC pantoate--beta-alanine liga... 45 4e-05
BP060613 34 0.066
TC18742 similar to UP|Q9FZG5 (Q9FZG5) T2E6.4, partial (3%) 33 0.15
TC14889 UP|Q7PI53 (Q7PI53) ENSANGP00000023856 (Fragment), partia... 32 0.25
AV780052 29 1.6
CN825238 29 2.1
BP055389 28 2.8
TC19943 similar to UP|AAN86570 (AAN86570) Trehalose-6-phosphate ... 27 6.2
TC10327 weakly similar to UP|NLT1_PRUDU (Q43017) Nonspecific lip... 27 8.1
>TC18710
Length = 843
Score = 83.2 bits (204), Expect = 1e-16
Identities = 39/93 (41%), Positives = 55/93 (58%)
Frame = +2
Query: 2 ILKVDMSKAYDRINWEYLRRVMLRLGFDAKWFGWIMMCVESVKYKVLVNQESEGPILPGS 61
I+K+DM+KAYDR+ W++L +L GF W IM+ V V Y +N +LP
Sbjct: 563 IIKLDMNKAYDRVEWKFLESSLLAFGFSTNWVKMIMILVSGVSYNYKINGVVGPKLLPQR 742
Query: 62 GLRKGDPLSPYLFILCAQGLTSLIDKAEGRGDL 94
GLR+GDP SPYLF+ + L+ LI + G+L
Sbjct: 743 GLRQGDPFSPYLFLFTMEVLSLLIQNSFNMGNL 841
>TC12836 GB|CAA71302.1|2292921|LJPANC pantoate--beta-alanine ligase {Lotus
corniculatus var. japonicus;} , complete
Length = 1310
Score = 44.7 bits (104), Expect = 4e-05
Identities = 22/43 (51%), Positives = 28/43 (64%)
Frame = +1
Query: 109 LLFVDDCFLFCRAT*REVNALKNILSIYEDASGQQINFQKSEI 151
LLFV D +FCRAT E N ++ IL +Y+ A G IN KSE+
Sbjct: 1102 LLFVGDGIIFCRATTPEENKIREILLMYQSAPGLLINLDKSEL 1230
>BP060613
Length = 378
Score = 33.9 bits (76), Expect = 0.066
Identities = 16/50 (32%), Positives = 30/50 (60%), Gaps = 1/50 (2%)
Frame = +1
Query: 305 LKARYYRRNAIFEANKGNNLSYVWKSI-HEAIPMVKQGTRWKIGNGDKIR 353
LKA Y+ + + K S+VW S+ H ++KQG +W++G+G+ ++
Sbjct: 220 LKALYFPHHDFLQTTKRTGASWVWSSLLHGRELLLKQG-QWQLGSGETVQ 366
>TC18742 similar to UP|Q9FZG5 (Q9FZG5) T2E6.4, partial (3%)
Length = 912
Score = 32.7 bits (73), Expect = 0.15
Identities = 15/46 (32%), Positives = 24/46 (51%)
Frame = -3
Query: 107 THLLFVDDCFLFCRAT*REVNALKNILSIYEDASGQQINFQKSEIF 152
+HL F D+ +F + + N+L I++ SG N KSE+F
Sbjct: 370 SHLCFADNLMVFSNGYLESIAIINNVLHIFQHLSGLTPNPAKSEVF 233
>TC14889 UP|Q7PI53 (Q7PI53) ENSANGP00000023856 (Fragment), partial (18%)
Length = 598
Score = 32.0 bits (71), Expect = 0.25
Identities = 20/62 (32%), Positives = 29/62 (46%), Gaps = 3/62 (4%)
Frame = +3
Query: 16 WEYLRRVMLRLGFDAKWFGWIMMCV---ESVKYKVLVNQESEGPILPGSGLRKGDPLSPY 72
W + ML +G KW W +CV E + ++ + Q + L G + DPLS Y
Sbjct: 36 WPF*MN*MLSIGSAGKWVVWECVCVTSLEKLMWRPMS*QSRDVTELSRFGRKCFDPLSIY 215
Query: 73 LF 74
LF
Sbjct: 216LF 221
>AV780052
Length = 584
Score = 29.3 bits (64), Expect = 1.6
Identities = 12/24 (50%), Positives = 14/24 (58%)
Frame = -3
Query: 90 GRGDLHGVRVCTGAPTFTHLLFVD 113
GRGDLH + V P THL + D
Sbjct: 201 GRGDLHRLEVINSMPPTTHLSYTD 130
>CN825238
Length = 692
Score = 28.9 bits (63), Expect = 2.1
Identities = 14/55 (25%), Positives = 26/55 (46%)
Frame = +3
Query: 383 LLQPGQKSWNLDVLQHYLSSQSVFEAMNTLLFNSTAYDERVWRPGKRGIYTVRSA 437
LL P + WN+ ++ ++ + L T + +WR + G ++VRSA
Sbjct: 297 LLIPNARRWNVQLIDQLFPKETAAVIKSIPLGWRTTRNSCLWRWSRNGAFSVRSA 461
>BP055389
Length = 535
Score = 28.5 bits (62), Expect = 2.8
Identities = 23/52 (44%), Positives = 29/52 (55%), Gaps = 1/52 (1%)
Frame = -1
Query: 378 LNVKELLQPGQKSW-NLDVLQHYLSSQSVFEAMNTLLFNSTAYDERVWRPGK 428
LNV +LQ KSW N+ + QHY S S+ E L+F ST RV+R K
Sbjct: 166 LNVS*VLQ**TKSWNNITMFQHY--SCSLIEENLQLMFRSTK-TTRVFRNAK 20
>TC19943 similar to UP|AAN86570 (AAN86570) Trehalose-6-phosphate
synthase/phosphatase (Fragment), partial (21%)
Length = 524
Score = 27.3 bits (59), Expect = 6.2
Identities = 17/37 (45%), Positives = 22/37 (58%), Gaps = 1/37 (2%)
Frame = +1
Query: 61 SGLRKGDPLSPYLFILCAQGLTSLIDKAEG-RGDLHG 96
S +R G L PY + +C QG +L DKA G GD +G
Sbjct: 46 SAVRDGMNLMPYEYTVCRQGSVAL-DKALGVEGDENG 153
>TC10327 weakly similar to UP|NLT1_PRUDU (Q43017) Nonspecific lipid-transfer
protein 1 precursor (LTP 1), partial (52%)
Length = 683
Score = 26.9 bits (58), Expect = 8.1
Identities = 17/54 (31%), Positives = 26/54 (47%), Gaps = 1/54 (1%)
Frame = -3
Query: 365 LAQPAA*YNRLLQLNVKELLQPGQKSWNLDVLQHYLSSQSVFEA-MNTLLFNST 417
L PA+ YN+ Q +PGQ+ + + +HY Q + M +LLF T
Sbjct: 654 LLSPASNYNKRTQA-----ARPGQRPYRHTIKEHYYKPQRLLHVHMCSLLFKHT 508
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.344 0.154 0.514
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,908,117
Number of Sequences: 28460
Number of extensions: 122704
Number of successful extensions: 1073
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1065
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1073
length of query: 520
length of database: 4,897,600
effective HSP length: 94
effective length of query: 426
effective length of database: 2,222,360
effective search space: 946725360
effective search space used: 946725360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC147202.9


