

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147179.8 + phase: 0
(307 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
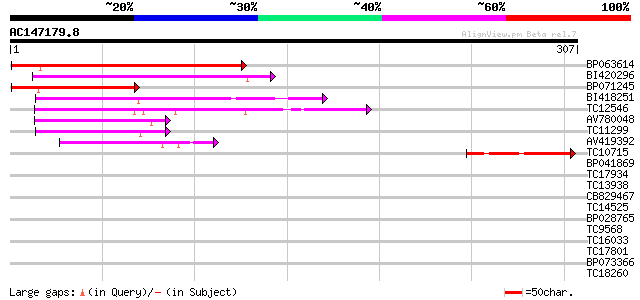
Score E
Sequences producing significant alignments: (bits) Value
BP063614 142 9e-35
BI420296 110 3e-25
BP071245 82 1e-16
BI418251 59 8e-10
TC12546 weakly similar to UP|Q9FJ30 (Q9FJ30) Similarity to heat ... 54 4e-08
AV780048 50 4e-07
TC11299 49 8e-07
AV419392 44 3e-05
TC10715 42 1e-04
BP041869 33 0.046
TC17934 similar to PIR|T06667|T06667 argininosuccinate synthase ... 33 0.046
TC13938 29 0.88
CB829467 28 2.0
TC14525 weakly similar to UP|CHMT_MEDSA (P93324) Isoliquiritigen... 28 2.0
BP028765 28 2.0
TC9568 28 2.5
TC16033 UP|Q7X9C1 (Q7X9C1) NIN-like protein 1, partial (88%) 28 2.5
TC17801 weakly similar to GB|AAO42978.1|28416895|BT004732 At3g13... 27 3.3
BP073366 27 3.3
TC18260 27 4.3
>BP063614
Length = 508
Score = 142 bits (357), Expect = 9e-35
Identities = 71/130 (54%), Positives = 92/130 (70%), Gaps = 3/130 (2%)
Frame = +2
Query: 2 KRGRKKHNDYVKED---RLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLI 58
KRGR+ N+ ED RLSDLP+C+LL ILS + A AV+TC +S+RW+ LWK LP+LI
Sbjct: 119 KRGRQSLNENENEDNKDRLSDLPDCILLHILSFVKAKAAVQTCILSTRWKDLWKRLPSLI 298
Query: 59 LCSSHFMRIKSFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKL 118
L SS+F KSF +FVSR+L LRD ST LH +DF G + P LLK++V YAVSHNV++L
Sbjct: 299 LLSSNFWTYKSFTKFVSRLLTLRDGSTALHGLDFEHDGHIQPHLLKRIVKYAVSHNVQRL 478
Query: 119 RVHACCDFQH 128
+ C +H
Sbjct: 479 GLSVTCAIEH 508
>BI420296
Length = 592
Score = 110 bits (275), Expect = 3e-25
Identities = 58/135 (42%), Positives = 82/135 (59%), Gaps = 3/135 (2%)
Frame = +2
Query: 13 KEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKSFNR 72
+ DRLSDLP+ VLL +L + +AV+TC +S+RW+ LWK + TL L SS F F+
Sbjct: 188 ERDRLSDLPDLVLLHVLKFMSTKQAVQTCVLSTRWKDLWKGVTTLALNSSDFATAPRFSE 367
Query: 73 FVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQ---HF 129
F+S +L R+ S LH +D +G V+P LL +V++YAVSH+V+ L + +
Sbjct: 368 FLSCVLSHRNDSVSLHNLDLRRKGCVEPELLNRVMSYAVSHDVQSLTIEFNLYLKLGFKL 547
Query: 130 SSCFFSCHTLTSLDL 144
C FSC TLT L L
Sbjct: 548 HPCIFSCRTLTYLKL 592
>BP071245
Length = 552
Score = 81.6 bits (200), Expect = 1e-16
Identities = 41/72 (56%), Positives = 52/72 (71%), Gaps = 3/72 (4%)
Frame = +1
Query: 2 KRGRKKHNDYVKE---DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLI 58
KRGR+ ++ E DRLSDLP+CVLL ILS ++A AV+TCT+S+RW+ LWK LP+LI
Sbjct: 334 KRGRESESESENEENKDRLSDLPDCVLLHILSFVNAKYAVQTCTLSTRWKDLWKRLPSLI 513
Query: 59 LCSSHFMRIKSF 70
L SS F K F
Sbjct: 514 LHSSDFSTFKGF 549
>BI418251
Length = 546
Score = 59.3 bits (142), Expect = 8e-10
Identities = 51/165 (30%), Positives = 72/165 (42%), Gaps = 7/165 (4%)
Frame = +1
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIK------ 68
DR+S LP+ VL +ILS L ++AV T + RW LW +PTL +++ K
Sbjct: 37 DRISKLPDEVLCQILSFLPTEDAVATSVLCKRWSSLWLSVPTLDFDDYRYLKGKKLKLQS 216
Query: 69 SFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKV-VTYAVSHNVEKLRVHACCDFQ 127
SF FV I+ R + P+ P KV + A+ VE L + C
Sbjct: 217 SFINFVYAIILSRALHQPIKNFTLSVISEECPYPDVKVWLNAAMQRQVENLDI--CLSLS 390
Query: 128 HFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLE 172
SC TL L L +D + ++LP+L TL LE
Sbjct: 391 TLPCSILSCTTLVVLKL------SDVKFHVFSCVDLPSLKTLHLE 507
>TC12546 weakly similar to UP|Q9FJ30 (Q9FJ30) Similarity to heat shock
transcription factor HSF30, partial (9%)
Length = 755
Score = 53.5 bits (127), Expect = 4e-08
Identities = 57/204 (27%), Positives = 84/204 (40%), Gaps = 21/204 (10%)
Frame = +2
Query: 14 EDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMR------- 66
ED +S LP+ +L ILS L + EAV T +S RW LW+ +PTL +++
Sbjct: 146 EDGISTLPDALLCHILSFLTSKEAVATSVLSRRWIPLWRSVPTLHFKDANYHTDIGHADH 325
Query: 67 --IKSFN----RFVSRILCLRDVSTPLH----AVDFLCRGVVDPRLLKKVVTYAVSHNVE 116
+K + V ++ RD P+ ++ +C+ DP + V V +E
Sbjct: 326 DIVKDVRSRHVQSVYAVILSRDFQLPIKKFYLRLNDVCQPFYDPANVSVWVNAVVQRQLE 505
Query: 117 KLRVHACCDF----QHFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLE 172
L + + S FSC TL L L G E FP S++ P L L L+
Sbjct: 506 HLDISLPYPMLSTPRANLSSIFSCRTLVVLKLRGGL----ELKRFP-SVHFPCLKVLHLQ 670
Query: 173 SFAFCVGDDGCAEPFSALNSLKNL 196
AE S L+NL
Sbjct: 671 GALLLHDVPYLAELLSGCPVLENL 742
>AV780048
Length = 501
Score = 50.4 bits (119), Expect = 4e-07
Identities = 30/78 (38%), Positives = 44/78 (55%), Gaps = 4/78 (5%)
Frame = +2
Query: 14 EDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSH-FMRIKSFNR 72
EDR S LP+ +L ILSLL EAV T +S RW LW+ +PTL S+ + + + R
Sbjct: 248 EDRFSSLPDPILCHILSLLTTKEAVTTSILSKRWIPLWRSVPTLDFDDSNWYGGDEVYAR 427
Query: 73 FVS---RILCLRDVSTPL 87
V +++ RD + P+
Sbjct: 428 LVEAVYKVILARDFNQPI 481
>TC11299
Length = 498
Score = 49.3 bits (116), Expect = 8e-07
Identities = 26/77 (33%), Positives = 39/77 (49%), Gaps = 4/77 (5%)
Frame = +1
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKS----F 70
DR+S LP+ ++ ILS L + AV T +S RW HLW+ + L S F
Sbjct: 85 DRISALPDEIICHILSFLPTENAVATAVLSKRWTHLWRSVSALDFSSVRMYEPDDHRFFF 264
Query: 71 NRFVSRILCLRDVSTPL 87
+ V +L R+ +TP+
Sbjct: 265 SDIVYSVLLFRNAATPI 315
>AV419392
Length = 318
Score = 43.9 bits (102), Expect = 3e-05
Identities = 31/105 (29%), Positives = 45/105 (42%), Gaps = 19/105 (18%)
Frame = +3
Query: 28 ILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKSFNRFVSRILCLR------ 81
ILS ++ +A++ +S RWR LW+ LP L S+ F SF FV +L
Sbjct: 6 ILSYVETKDAMQCSVLSKRWRSLWRALPVLNFDSASFTHYSSFANFVLHVLHSSTSLTLT 185
Query: 82 ------DVSTPLHAV-------DFLCRGVVDPRLLKKVVTYAVSH 113
D+S P+ LC G+V + V T+ V H
Sbjct: 186 HCKFWFDISNPIFCCLQICIDRQDLCEGLV-RMFIHHVTTHGVQH 317
>TC10715
Length = 534
Score = 42.0 bits (97), Expect = 1e-04
Identities = 22/59 (37%), Positives = 37/59 (62%)
Frame = +3
Query: 248 GIPVLKLCRSKINLSSVKHVNIEVRMWTDYADASLVLLNWLAELANIKSLTLNHTALKV 306
G P +C N+SS++HV+ ++ + S +LL+WL +L NIKSLT++ + L+V
Sbjct: 3 GTPFETICGR--NISSIEHVHFDIHWQCE--KYSFILLSWLVDLTNIKSLTVSTSTLQV 167
>BP041869
Length = 257
Score = 33.5 bits (75), Expect = 0.046
Identities = 20/61 (32%), Positives = 31/61 (50%)
Frame = +1
Query: 158 PKSLNLPALTTLSLESFAFCVGDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLT 217
P S+NL L ++ F F D C L +L+ L+C+VL +C+S+V L+
Sbjct: 13 PSSINLFLLVSIPPPIFDFF---DQCNTALLLFLLLSSLLSLFCSVLSLRVVCLSTVDLS 183
Query: 218 N 218
N
Sbjct: 184 N 186
>TC17934 similar to PIR|T06667|T06667 argininosuccinate synthase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (11%)
Length = 564
Score = 33.5 bits (75), Expect = 0.046
Identities = 14/35 (40%), Positives = 23/35 (65%)
Frame = -1
Query: 17 LSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLW 51
++ LP+ + ILS L +EA KT +S +WR++W
Sbjct: 564 INQLPDGIPGAILSKLPINEAAKTSILSRKWRYMW 460
>TC13938
Length = 584
Score = 29.3 bits (64), Expect = 0.88
Identities = 20/59 (33%), Positives = 30/59 (49%), Gaps = 4/59 (6%)
Frame = -1
Query: 182 GCAEPFSALNSLKNLMILYC---NVLDAESLCISSVTLTNLTIVGDP-VYYSTVELSTP 236
G A PFS +SL+ + YC N+L +C++S NL +V + Y + L TP
Sbjct: 227 GFAAPFSCQSSLRKQALCYCE*GNILFCF*ICLTSYVHINLFLVPNK*NYIRFLPLMTP 51
>CB829467
Length = 567
Score = 28.1 bits (61), Expect = 2.0
Identities = 21/81 (25%), Positives = 39/81 (47%), Gaps = 3/81 (3%)
Frame = +2
Query: 167 TTLSLESFAFCVGDDGCAEPFSALNSLKNLMI---LYCNVLDAESLCISSVTLTNLTIVG 223
TTL L S C + +PF +LK+L + + + L IS+ L+NLT++
Sbjct: 2 TTLHLNSLTLCYTGNEVLDPFVNCVNLKDLHLSEMSFKSQLIPRDFVISAPRLSNLTLMC 181
Query: 224 DPVYYSTVELSTPSLFTFDFV 244
+ + + ++ P L F ++
Sbjct: 182 NR-FKCKLVVNAPQLINFSYL 241
>TC14525 weakly similar to UP|CHMT_MEDSA (P93324) Isoliquiritigenin
2'-O-methyltransferase (Chalcone O-methyltransferase)
(ChOMT) , partial (58%)
Length = 1077
Score = 28.1 bits (61), Expect = 2.0
Identities = 11/33 (33%), Positives = 17/33 (51%)
Frame = -3
Query: 47 WRHLWKHLPTLILCSSHFMRIKSFNRFVSRILC 79
W HLW++ T L H + F +++ R LC
Sbjct: 922 WHHLWEYFQTCFL--QHAQSLNEF*KYLDRRLC 830
>BP028765
Length = 529
Score = 28.1 bits (61), Expect = 2.0
Identities = 25/73 (34%), Positives = 35/73 (47%), Gaps = 6/73 (8%)
Frame = +3
Query: 20 LPNCVLLRILSLLDADEAVKTCTISSRWRHLW---KHL-PTLILCSSHFMRIKSFN--RF 73
L NC+ ++LS V+T T +S WR W KHL P L S + F R+
Sbjct: 303 LGNCIPHKLLSSRRKRAHVRTSTSTSNWRCQWGLMKHLCPRPQLPCSLPRK*VCFRIPRW 482
Query: 74 VSRILCLRDVSTP 86
R + +R +STP
Sbjct: 483 SRRNIVMRSISTP 521
>TC9568
Length = 709
Score = 27.7 bits (60), Expect = 2.5
Identities = 11/37 (29%), Positives = 18/37 (47%)
Frame = +1
Query: 113 HNVEKLRVHACCDFQHFSSCFFSCHTLTSLDLCVGPP 149
H +LR+ +CC + F + T T + C+G P
Sbjct: 505 HLFTELRIFSCCSSSVYPFIFLTALTRTHIQSCLGVP 615
>TC16033 UP|Q7X9C1 (Q7X9C1) NIN-like protein 1, partial (88%)
Length = 2554
Score = 27.7 bits (60), Expect = 2.5
Identities = 12/22 (54%), Positives = 18/22 (81%)
Frame = +2
Query: 7 KHNDYVKEDRLSDLPNCVLLRI 28
+HN YVK+DRLS + +C ++RI
Sbjct: 935 RHN-YVKQDRLSTVSSCQIVRI 997
>TC17801 weakly similar to GB|AAO42978.1|28416895|BT004732 At3g13950
{Arabidopsis thaliana;}, partial (22%)
Length = 423
Score = 27.3 bits (59), Expect = 3.3
Identities = 13/41 (31%), Positives = 25/41 (60%), Gaps = 3/41 (7%)
Frame = -3
Query: 143 DLCVGPPRTDERLSFPKSLN---LPALTTLSLESFAFCVGD 180
++C+ P + + SFP++LN +P ++SL + A C+ D
Sbjct: 346 EICISVPVHNTQHSFPRALNGIIVPTNPSISLWNTAICLKD 224
>BP073366
Length = 290
Score = 27.3 bits (59), Expect = 3.3
Identities = 14/46 (30%), Positives = 25/46 (53%)
Frame = -3
Query: 186 PFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYSTV 231
PF NS+ +++ + +L ESLC ++L + + P+Y TV
Sbjct: 180 PFQIKNSIMLILVCFHYILFMESLCA*IISLVDFLLFA-PMYIRTV 46
>TC18260
Length = 727
Score = 26.9 bits (58), Expect = 4.3
Identities = 14/37 (37%), Positives = 21/37 (55%)
Frame = -3
Query: 228 YSTVELSTPSLFTFDFVCYEGIPVLKLCRSKINLSSV 264
Y+T+ T F+F F Y + +LK+ S+ NL SV
Sbjct: 335 YNTLTCGTD*SFSFSFQEYNQLNILKIHASQTNLLSV 225
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.138 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,927,764
Number of Sequences: 28460
Number of extensions: 115703
Number of successful extensions: 940
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 930
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 939
length of query: 307
length of database: 4,897,600
effective HSP length: 90
effective length of query: 217
effective length of database: 2,336,200
effective search space: 506955400
effective search space used: 506955400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC147179.8


