

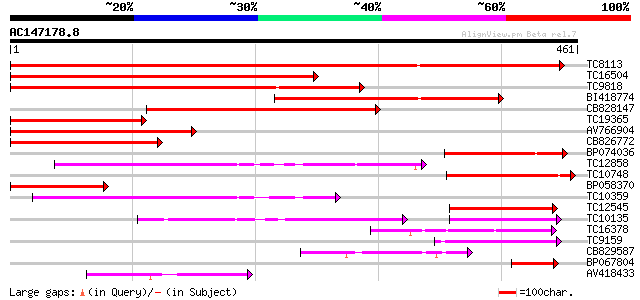
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.8 + phase: 0
(461 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter li... 499 e-142
TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporte... 347 2e-96
TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, ... 292 9e-80
BI418774 265 9e-72
CB828147 242 1e-64
TC19365 weakly similar to UP|O23213 (O23213) Sugar transporter l... 178 2e-45
AV766904 175 1e-44
CB826772 161 2e-40
BP074036 142 9e-35
TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter,... 131 3e-31
TC10748 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter,... 117 4e-27
BP058370 106 9e-24
TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partia... 106 9e-24
TC12545 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter,... 105 1e-23
TC10135 89 2e-18
TC16378 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22... 70 7e-13
TC9159 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose tr... 68 4e-12
CB829587 67 5e-12
BP067804 67 6e-12
AV418433 61 3e-10
>TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (93%)
Length = 1865
Score = 499 bits (1284), Expect = e-142
Identities = 246/451 (54%), Positives = 325/451 (71%)
Frame = +2
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GALLFIKE++ ISD Q ++LAGILN CAL C+ AG+ SDYIGRRYTI L+SI F +G
Sbjct: 179 MSGALLFIKEDIGISDTQQEVLAGILNICALVGCLAAGKTSDYIGRRYTIFLASILFLVG 358
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++ MGYG +F ILM GRC+ G GVGFAL VYSAE+SS S RGFLTSLP++ I +G
Sbjct: 359 AVFMGYGPNFAILMFGRCVCGLGVGFALTTAPVYSAELSSASTRGFLTSLPEVCIGLGIF 538
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
+GY+SNYFLGKL+L LGWR+ML + +IPS+GL + +L + ESPRWLVMQGRLG AKKVLL
Sbjct: 539 IGYISNYFLGKLALTLGWRLMLGLAAIPSLGLALGILTMPESPRWLVMQGRLGCAKKVLL 718
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
+SN+ +EAE R ++I + G DE C V +K+ G G KE+F +P+P V R+LI
Sbjct: 719 EVSNTTEEAELRFRDIIVSAGFDEKCNDEFVKQPQKSHHGEGVWKELFLRPTPPVRRMLI 898
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AA+G+H F++ G+E + LY PRIF + G+T K LLLAT+G G+++ F +S FLLD+
Sbjct: 899 AAVGIHFFEHATGIEAVMLYGPRIFKKAGVTTKDRLLLATIGTGLTKITFLTISTFLLDR 1078
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GRR LL +S G+IF + L +VE S E+ +WA+ +I+ Y F +G+
Sbjct: 1079VGRRRLLQISVAGMIFGLTILGFSLTMVEYS--SEKLVWALSLSIVATYTYVAFFNVGLA 1252
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
VTWVYS+EIFPLRLRAQG + V +NR N A+ SFISIYK IT+GG FFLF G +V+
Sbjct: 1253PVTWVYSSEIFPLRLRAQGNSIGVAVNRGMNAAISMSFISIYKAITIGGAFFLFAGMSVV 1432
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGKNSNSE 451
W F+Y LPETKG++LE+ME +F K S+ +
Sbjct: 1433AWVFFYFCLPETKGKALEEMEMVFSKKSDDK 1525
>TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporter, partial
(37%)
Length = 903
Score = 347 bits (891), Expect = 2e-96
Identities = 167/251 (66%), Positives = 209/251 (82%)
Frame = +2
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M G LLFIKE+L++SD+QVQ G+L+ ALPACM AGR++DYIGRRYTI+L++I F +G
Sbjct: 149 MTGVLLFIKEDLQLSDLQVQFSVGLLHMSALPACMTAGRIADYIGRRYTIILTAIIFLVG 328
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
S+LMGYG S+PILMIG I+G G+GF+LI+ VY AEIS PS+RGFLTS ++S N G
Sbjct: 329 SVLMGYGPSYPILMIGLSISGIGLGFSLIVAPVYCAEISPPSHRGFLTSFLEVSTNAGIA 508
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNYF GKLS+RLGWR+ML + ++PS+ LV LML VESPRWLVMQGR+G+A+KVLL
Sbjct: 509 LGYVSNYFFGKLSIRLGWRLMLVVHAVPSLALVALMLNWVESPRWLVMQGRVGEARKVLL 688
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
L+SN+K+EAEQR+KEIK AVGIDE CTQ+IV V +TR+GGGA +EMF KP+P V RILI
Sbjct: 689 LVSNTKEEAEQRLKEIKVAVGIDEKCTQDIVQVPNRTRNGGGAFREMFCKPTPPVRRILI 868
Query: 241 AAIGVHIFQNI 251
AA+GVH FQ +
Sbjct: 869 AAVGVHAFQQL 901
>TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 1162
Score = 292 bits (747), Expect = 9e-80
Identities = 143/288 (49%), Positives = 209/288 (71%)
Frame = +2
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA ++IK +L++SD+++++L GI+N +L +AGR SD+IGRRYTI+ + FF+G
Sbjct: 299 MSGAAIYIKRDLKVSDVKIEILLGIINLYSLIGSGLAGRTSDWIGRRYTIVFAGAIFFVG 478
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG+ ++ LM GR IAG G+G+AL+I VY+AE+S S RGFLTS P++ IN G L
Sbjct: 479 ALLMGFSPNYWFLMFGRFIAGIGIGYALMIAPVYTAEVSPASSRGFLTSFPEVFINGGIL 658
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SN+ KLSL++GWR+ML + ++PS+ L + +L + ESPRWLVM+GRLGDA KVL
Sbjct: 659 LGYISNFAFSKLSLKVGWRMMLGVGALPSVILGVGVLAMPESPRWLVMRGRLGDAIKVLN 838
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+S +EA+ R+ +IK A GI E+CT ++V VSK++ +G G KE+F P+P + I+I
Sbjct: 839 KTSDSPEEAQLRLADIKRAAGIPESCTDDVVEVSKRS-TGEGVWKELFLYPTPAIRHIVI 1015
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQT 288
AA+G+H FQ G++ + LYSP IF + GI LLATV +G +T
Sbjct: 1016AALGIHFFQQASGIDAVVLYSPTIFEKAGIKSDTDKLLATVAVGFVKT 1159
>BI418774
Length = 554
Score = 265 bits (678), Expect = 9e-72
Identities = 131/186 (70%), Positives = 156/186 (83%)
Frame = +3
Query: 216 KTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGT 275
KTR+G GAL+E+F KPSP V RILIAAIGVH+FQ ICG+EGI +YSPR+F + GIT K
Sbjct: 3 KTRNGEGALRELFVKPSPPVRRILIAAIGVHVFQQICGIEGICIYSPRVFEKTGITSKSN 182
Query: 276 LLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGE 335
LLLATVG+GISQ +FT +S FLLD++GRRILLL+SSGGV+ ++LGL CS I++ SK E
Sbjct: 183 LLLATVGLGISQAVFTFISAFLLDRVGRRILLLISSGGVVVTLLGLSFCSFIMQQSK--E 356
Query: 336 EPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVV 395
EPLW I FTI+ IYI F AIGIG VTWVYS+EIFPLRLRAQGLGVCV++NR+ NVAV+
Sbjct: 357 EPLWGISFTIVAIYIFVAFVAIGIGPVTWVYSSEIFPLRLRAQGLGVCVLVNRLANVAVL 536
Query: 396 TSFISI 401
TSFISI
Sbjct: 537 TSFISI 554
>CB828147
Length = 570
Score = 242 bits (617), Expect = 1e-64
Identities = 116/190 (61%), Positives = 150/190 (78%)
Frame = +1
Query: 112 DLSINIGFLLGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGR 171
+ SINIG LLGY+S +F KL L LGWR+MLA+P+ PSI L+ LML+L ESPRWL+MQGR
Sbjct: 1 EFSINIGILLGYMSTFFFEKLRLNLGWRMMLAVPAAPSIALIFLMLKLGESPRWLIMQGR 180
Query: 172 LGDAKKVLLLISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKP 231
+GDAKKVLLL+ N+KQEAEQR+ EIK AVGI+ N TQ IV V KKTR GGGALKE+F KP
Sbjct: 181 VGDAKKVLLLVFNTKQEAEQRLAEIKTAVGIEPNNTQEIVQVPKKTRHGGGALKELFCKP 360
Query: 232 SPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFT 291
+P V RI++AA+G+H+F ++ G+ I LYSPRIF + GIT K LLL T+G+G+ + +F+
Sbjct: 361 TPLVRRIMVAAVGLHVFMHLGGIGVILLYSPRIFEKTGITSKSKLLLCTIGMGVMKLVFS 540
Query: 292 LLSCFLLDKI 301
+S F +D +
Sbjct: 541 FISIFFMDGV 570
>TC19365 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (24%)
Length = 441
Score = 178 bits (451), Expect = 2e-45
Identities = 89/111 (80%), Positives = 95/111 (85%)
Frame = +2
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
MAGALLFIKEEL I+DMQVQLL GILN CALPACM+AGR SDY+GRRYTI+LS++ LG
Sbjct: 107 MAGALLFIKEELGINDMQVQLLGGILNVCALPACMVAGRTSDYVGRRYTIILSAVILLLG 286
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLP 111
SILMGYG SFPILMIGRC AG GVG ALII VYS EIS PS RGFLTSLP
Sbjct: 287 SILMGYGPSFPILMIGRCTAGLGVGLALIIAPVYSTEISLPSNRGFLTSLP 439
>AV766904
Length = 611
Score = 175 bits (444), Expect = 1e-44
Identities = 83/152 (54%), Positives = 117/152 (76%)
Frame = +3
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA+++IK +L+ISD+Q+++L+GI+N + IAGR SD+IGRRYTI+L+ + FF+G
Sbjct: 153 MSGAVIYIKRDLKISDVQIEILSGIMNIYSPIGSYIAGRTSDWIGRRYTIVLAGLIFFVG 332
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
+ILMG+ ++ LM GR +AG G+GFA +I VY++EIS RGFLTSLP++ +N G L
Sbjct: 333 AILMGFSPNYAFLMFGRFVAGVGIGFAFLIAPVYTSEISPTLSRGFLTSLPEVFLNAGIL 512
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGL 152
+GY+SNY KL LRLGWR+ML I +IPSI L
Sbjct: 513 IGYISNYTFSKLPLRLGWRLMLGIGAIPSIFL 608
>CB826772
Length = 490
Score = 161 bits (408), Expect = 2e-40
Identities = 81/124 (65%), Positives = 96/124 (77%)
Frame = +1
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M GALLFIKE+L+ISD+Q+Q L GI + C LPA M AGR+SDYIGRRYTI+L+SI F G
Sbjct: 118 MTGALLFIKEDLQISDLQLQFLVGIFHMCGLPASMAAGRISDYIGRRYTIILASIAFLSG 297
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
SIL GY S+ ILMIG I+G G GF LI+ +Y AEIS PS+RG LTSL + SINIG L
Sbjct: 298 SILTGYSPSYLILMIGNFISGIGAGFTLIVAPLYCAEISPPSHRGSLTSLTEFSINIGIL 477
Query: 121 LGYL 124
LGY+
Sbjct: 478 LGYM 489
>BP074036
Length = 405
Score = 142 bits (359), Expect = 9e-35
Identities = 68/100 (68%), Positives = 82/100 (82%)
Frame = -2
Query: 354 FNAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFL 413
F +IGIG VTW+YS+EIFP+ LRAQGL VCV +NRI N+A++TSFISIYK IT+GG F
Sbjct: 404 FMSIGIGPVTWIYSSEIFPITLRAQGLAVCVAVNRIVNMAMLTSFISIYKAITMGGCLFA 225
Query: 414 FVGTNVLGWWFYYSFLPETKGRSLEDMETIFGKNSNSEVQ 453
G NVL +WFY++ LPETKGRSLEDME +FGK S +EVQ
Sbjct: 224 LAGVNVLAFWFYFT-LPETKGRSLEDMEIVFGKVSKTEVQ 108
>TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter, partial
(51%)
Length = 882
Score = 131 bits (329), Expect = 3e-31
Identities = 89/305 (29%), Positives = 152/305 (49%), Gaps = 2/305 (0%)
Frame = +3
Query: 37 AGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSA 96
A ++ +G R T+++ +FF G++ G +L++GR + GFG+G A V +Y +
Sbjct: 12 ASTITRIMGMRATMIVGGLFFVSGALFNGLADGIWMLIVGRLLLGFGIGCANQSVPIYLS 191
Query: 97 EISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILM 156
E++ YRG L LSI IG + L NY+ K+ GWR+ L + ++P++ V+
Sbjct: 192 EMAPYKYRGGLNMCFQLSITIGIFVANLFNYYFAKILNGQGWRLSLGLGAVPAVIFVVGS 371
Query: 157 LQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKK 216
L L +SP LV +GR A++ L+ I + + E +K+I A +N+ H K
Sbjct: 372 LCLPDSPSSLVARGRHEAARQELVKIRGT-TDIEAELKDIITA----SEALENVKHPWK- 533
Query: 217 TRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTL 276
L E Y+P L+ A+ + FQ G+ I Y+P +F +G +L
Sbjct: 534 ------TLLERKYRPQ------LVFAVCIPFFQQFTGLNVITFYAPILFRTIGFGPTASL 677
Query: 277 LLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCVCSAIV--ENSKLG 334
+ A + IG + + TL+S F++DK GRR L L ++ + + + A+ + G
Sbjct: 678 MSAVI-IGSFKPVSTLISIFVVDKFGRRTLFLEGGAQMLICQIIMTIAIAVTFGTSGNPG 854
Query: 335 EEPLW 339
+ P W
Sbjct: 855 QLPKW 869
>TC10748 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(21%)
Length = 550
Score = 117 bits (293), Expect = 4e-27
Identities = 55/105 (52%), Positives = 78/105 (73%)
Frame = +2
Query: 356 AIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFV 415
+IG G +TWVYS+EIFPLRLRAQG + V++NR+T+ + +F+S+ K IT+GG FFLF
Sbjct: 29 SIGAGPITWVYSSEIFPLRLRAQGCAMGVVVNRVTSGVISMTFLSLSKGITIGGAFFLFG 208
Query: 416 GTNVLGWWFYYSFLPETKGRSLEDMETIFGKNSNSEVQMKYGSNN 460
G + GW F+Y+ LPET+G++LEDME FG+ S S+++ G N
Sbjct: 209 GIAICGWIFFYTMLPETRGKTLEDMEGSFGQ-SKSKIKGNKGVGN 340
>BP058370
Length = 358
Score = 106 bits (264), Expect = 9e-24
Identities = 53/80 (66%), Positives = 63/80 (78%)
Frame = +3
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M GALLFIKE+L+ISD+Q+Q L GI + C LPA M AGR+SDYIGRRYTI+L+SI F G
Sbjct: 117 MTGALLFIKEDLQISDLQLQFLVGIFHMCGLPASMAAGRISDYIGRRYTIILASIAFLSG 296
Query: 61 SILMGYGSSFPILMIGRCIA 80
SIL GY S+ ILMIG I+
Sbjct: 297 SILTGYSPSYLILMIGNFIS 356
>TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partial (90%)
Length = 1099
Score = 106 bits (264), Expect = 9e-24
Identities = 72/251 (28%), Positives = 118/251 (46%)
Frame = +2
Query: 19 VQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRC 78
+QL L L + A + +GRR T++++ +FF G + + +L++GR
Sbjct: 389 LQLFTSSLYLAGLVSTFFASYTTRTLGRRMTMLIAGVFFIAGVVFNVTAQNLAMLIVGRI 568
Query: 79 IAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGW 138
+ G GVGFA V ++ +EI+ RG L L L+I IG L L NY K+ GW
Sbjct: 569 LLGCGVGFANQAVPLFLSEIAPSRIRGALNILFQLNITIGILFANLINYGTNKIKGGWGW 748
Query: 139 RIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKN 198
R+ L + IP++ L + L +V++P L+ +GR + K VL I + E E+
Sbjct: 749 RLSLGLAGIPAVLLTVGALLVVDTPNSLIERGRTEEGKAVLKKIRGT-DNVEPEFLELVE 925
Query: 199 AVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIF 258
A V+K+ + L + +P +I +I + IFQ G+ I
Sbjct: 926 A-----------SRVAKEVKDPFRNLLKRRNRPQ------IIISIALQIFQQFTGINAIM 1054
Query: 259 LYSPRIFGRMG 269
Y+P +F +G
Sbjct: 1055FYAPVLFNTLG 1087
>TC12545 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(18%)
Length = 482
Score = 105 bits (263), Expect = 1e-23
Identities = 46/88 (52%), Positives = 65/88 (73%)
Frame = +2
Query: 358 GIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGT 417
G G +TWVYS+EIFPLRLRAQG+ + ++NR+T + +F+S+ IT+GG FFLF G
Sbjct: 2 GSGPITWVYSSEIFPLRLRAQGVAIGAVVNRLTRGVISMTFLSLSNAITIGGAFFLFAGI 181
Query: 418 NVLGWWFYYSFLPETKGRSLEDMETIFG 445
V W F+Y+ LPET+G++LE++E FG
Sbjct: 182 AVCAWIFHYTLLPETRGQTLEEIEGSFG 265
>TC10135
Length = 1501
Score = 89.0 bits (219), Expect = 2e-18
Identities = 67/219 (30%), Positives = 106/219 (47%)
Frame = +2
Query: 105 GFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPR 164
G L SL I G L YL N L + WR ML + + P+I ++LML L ESPR
Sbjct: 2 GALVSLNSFLITGGQFLSYLIN--LAFTNAPGTWRWMLGVAAAPAIIQIVLMLSLPESPR 175
Query: 165 WLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGAL 224
WL +GR ++K +L I + ++ + ++ +K +V ++ + SK ++ +
Sbjct: 176 WLYRKGREEESKSILKKIY-APEDVDAEIEALKESV-------ESEIEESKTSK-----I 316
Query: 225 KEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIG 284
M + V R L A +G+ FQ G+ + YSP I G T LL ++
Sbjct: 317 SMMTLLKTTTVRRGLYAGMGLQFFQQFVGINTVMYYSPTIVQLAGFASNRTALLLSLITS 496
Query: 285 ISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCV 323
++LS + +DK GR+ L L+S GV+ S+ L V
Sbjct: 497 GLNAFGSILSIYFIDKTGRKKLALISLCGVVLSLALLTV 613
Score = 60.8 bits (146), Expect = 4e-10
Identities = 33/91 (36%), Positives = 52/91 (56%)
Frame = +2
Query: 358 GIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGT 417
G+G V WV ++EI+P R R G+ I+N+ V SF+S+ +TI TF +F
Sbjct: 935 GMGTVPWVINSEIYPFRYRGVCGGIASTTVWISNLIVSQSFLSLTQTIGTAWTFMMFGIL 1114
Query: 418 NVLGWWFYYSFLPETKGRSLEDMETIFGKNS 448
V+ +F F+PETKG +E++E + + S
Sbjct: 1115 AVVAIFFVIVFVPETKGVPMEEVEKMLEQRS 1207
>TC16378 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;}, partial (31%)
Length = 738
Score = 70.1 bits (170), Expect = 7e-13
Identities = 45/155 (29%), Positives = 88/155 (56%), Gaps = 4/155 (2%)
Frame = +2
Query: 294 SCFLLDKIGRRILLLVSSGGVIFSMLGLCVC----SAIVENSKLGEEPLWAIIFTIIVIY 349
S +L+DK GRR+LLL+SS + S++ + + + E+S L + + I+ + ++
Sbjct: 2 STWLVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYK--ILGIVSVVGLVV 175
Query: 350 IMAGFNAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGG 409
++ GF+ +G+G + WV +EI P+ ++ + N +T+ A+ T ++ T + GG
Sbjct: 176 MVIGFS-LGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAI-TMTANLLLTWSSGG 349
Query: 410 TFFLFVGTNVLGWWFYYSFLPETKGRSLEDMETIF 444
TF ++ F ++PETKGR+LE++++ F
Sbjct: 350 TFTMYTVVAAFTVVFTALWVPETKGRTLEEIQSSF 454
>TC9159 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose transporter
HXT4 (Low-affinity glucose transporter LGT1), partial
(5%)
Length = 728
Score = 67.8 bits (164), Expect = 4e-12
Identities = 36/103 (34%), Positives = 62/103 (59%)
Frame = +3
Query: 346 IVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTI 405
+ +YI+ F + G+G V WV ++EI+PLR R G+ ++N+ V SF+S+ +TI
Sbjct: 132 LALYII--FFSPGMGTVPWVVNSEIYPLRYRGICGGIASTTVWVSNLVVSQSFLSLTQTI 305
Query: 406 TLGGTFFLFVGTNVLGWWFYYSFLPETKGRSLEDMETIFGKNS 448
TF +F ++G +F F+PETKG +E++E++ K +
Sbjct: 306 GTAWTFMMFGIIAIIGIFFVLIFVPETKGVPMEEVESMLNKRA 434
>CB829587
Length = 548
Score = 67.4 bits (163), Expect = 5e-12
Identities = 45/144 (31%), Positives = 72/144 (49%), Gaps = 4/144 (2%)
Frame = +2
Query: 237 RILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITD--KGTLLLATVGIGISQTLFTLLS 294
R ++ +G+ + Q G+ GI Y+ F G++ GT+ A + Q FT+L
Sbjct: 131 RSVVIGVGLMVCQQSVGINGIGFYTAETFVAAGVSSGKAGTIAYACI-----QVPFTILG 295
Query: 295 CFLLDKIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTI--IVIYIMA 352
L+DK GRR L+ VSSGG + + + L E W I ++IYI A
Sbjct: 296 AILMDKSGRRPLITVSSGGTFLGCFITGIAFFLKDQGLLLE---WVPTLAIGGVLIYIAA 466
Query: 353 GFNAIGIGAVTWVYSTEIFPLRLR 376
+IG+G+V WV +E+FP+ ++
Sbjct: 467 --YSIGLGSVPWVIMSEVFPIHIK 532
>BP067804
Length = 412
Score = 67.0 bits (162), Expect = 6e-12
Identities = 29/38 (76%), Positives = 32/38 (83%)
Frame = -2
Query: 409 GTFFLFVGTNVLGWWFYYSFLPETKGRSLEDMETIFGK 446
GTFF+ G N++ W FYY FLPETKGRSLEDMETIFGK
Sbjct: 408 GTFFMLAGINLVAWSFYYFFLPETKGRSLEDMETIFGK 295
>AV418433
Length = 401
Score = 61.2 bits (147), Expect = 3e-10
Identities = 41/140 (29%), Positives = 70/140 (49%), Gaps = 5/140 (3%)
Frame = +3
Query: 63 LMGYGSSFPILM-IGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDL----SINI 117
L+ Y S P+L+ IGR G+G+G +V V+ AEI+ RG LT+L ++++
Sbjct: 15 LVIYFSQGPVLLDIGRVATGYGMGVFSYVVPVFIAEIAPKELRGALTTLNQFMLVSAMSV 194
Query: 118 GFLLGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKK 177
F++G + L WR + IP+ L++ + + ESPRWL +G D +
Sbjct: 195 SFVIGNV-----------LSWRALALTGLIPTAVLLLGLFFIPESPRWLAKRGCREDFEA 341
Query: 178 VLLLISNSKQEAEQRMKEIK 197
L ++ + Q +EI+
Sbjct: 342 ALQILRGKDADISQEAEEIQ 401
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.327 0.144 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,085,573
Number of Sequences: 28460
Number of extensions: 118457
Number of successful extensions: 823
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 800
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 809
length of query: 461
length of database: 4,897,600
effective HSP length: 93
effective length of query: 368
effective length of database: 2,250,820
effective search space: 828301760
effective search space used: 828301760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC147178.8


