

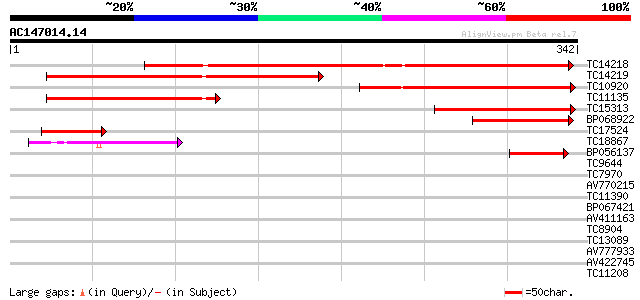
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147014.14 + phase: 0 /pseudo
(342 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14218 similar to UP|Q42896 (Q42896) Fructokinase , partial (77%) 266 5e-72
TC14219 homologue to UP|SCRK_SOLTU (P37829) Fructokinase , part... 203 3e-53
TC10920 similar to UP|O04897 (O04897) Fructokinase , partial (35%) 164 3e-41
TC11135 homologue to UP|SCRK_SOLTU (P37829) Fructokinase , part... 123 5e-29
TC15313 similar to UP|Q9FLH8 (Q9FLH8) Fructokinase 1, partial (23%) 107 4e-24
BP068922 63 6e-11
TC17524 similar to UP|O04897 (O04897) Fructokinase , partial (12%) 59 1e-09
TC18867 similar to UP|Q9ASV2 (Q9ASV2) At1g06730/F4H5_22, partial... 46 1e-05
BP056137 44 5e-05
TC9644 homologue to UP|Q9M394 (Q9M394) Fructokinase-like protein... 37 0.005
TC7970 similar to UP|ADK2_ARATH (Q9LZG0) Adenosine kinase 2 (AK... 36 0.011
AV770215 31 0.030
TC11390 similar to UP|Q8L993 (Q8L993) Carbohydrate kinase-like p... 34 0.031
BP067421 33 0.053
AV411163 32 0.12
TC8904 homologue to UP|G3PB_PEA (P12859) Glyceraldehyde 3-phosph... 31 0.26
TC13089 31 0.26
AV777933 30 0.59
AV422745 30 0.77
TC11208 weakly similar to GB|AAM78055.1|21928053|AY125545 At4g29... 30 0.77
>TC14218 similar to UP|Q42896 (Q42896) Fructokinase , partial (77%)
Length = 1170
Score = 266 bits (679), Expect = 5e-72
Identities = 141/260 (54%), Positives = 185/260 (70%), Gaps = 1/260 (0%)
Frame = +1
Query: 82 FGHMLSDILKQ-NGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRSE 140
FG ML D + +GV G+ FDE ARTALAF +L+ DG+ EFMFYRNPSAD+L + E
Sbjct: 109 FGEMLIDFVPTVSGVRADGITFDEGARTALAFVTLRA--DGEREFMFYRNPSADMLLKPE 282
Query: 141 EIDKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTEAA 200
E++ LI+ A +FHYGS+SLI EP RS H++A+ AK G +LSY PNL +PLWPS + A
Sbjct: 283 ELNLELIRSAKVFHYGSISLIVEPCRSAHLKAMEVAKDAGCLLSYDPNLRLPLWPSADEA 462
Query: 201 REGIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYY 260
R+ I+SIW+ AD+IKVS E+ LT G+D DD+ + L+H NLKLLLVT G G +YY
Sbjct: 463 RKQILSIWDKADLIKVSDNELEFLT-GSDKIDDESAL-SLWHPNLKLLLVTLGEHGSKYY 636
Query: 261 TKDFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAAT 320
TK+F+G V G+ V +DTTGAGDSFVG LS + + I +DE LRE L ++NACGA T
Sbjct: 637 TKNFRGTVEGYHVNTVDTTGAGDSFVGSLLSKIVDDQSILEDEPRLREVLKYSNACGAIT 816
Query: 321 VTGRGAIPSLPTKSSVLRVM 340
T +GAIP+LP + VL+++
Sbjct: 817 TTKKGAIPALPKEEDVLKLI 876
>TC14219 homologue to UP|SCRK_SOLTU (P37829) Fructokinase , partial (53%)
Length = 655
Score = 203 bits (517), Expect = 3e-53
Identities = 99/167 (59%), Positives = 132/167 (78%)
Frame = +1
Query: 23 LVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGNDEF 82
L+V FGEM+I+ VPT+ GVSL++A + K+P GA A V++A+SRLGG++AF+GK+G+DEF
Sbjct: 154 LIVSFGEMLIDFVPTVSGVSLAEAPGFLKAPGGAPANVAIAVSRLGGNAAFVGKLGDDEF 333
Query: 83 GHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRSEEI 142
GHML+ ILK+NGV G+ FDE ARTALAF +L+ DG+ EFMFYRNPSAD+L + EE+
Sbjct: 334 GHMLAGILKENGVRADGITFDEGARTALAFVTLR--ADGEREFMFYRNPSADMLLKPEEL 507
Query: 143 DKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNL 189
+ LI+ A +FHYGS+SLI EP RS H++A+ AK G +LSY PNL
Sbjct: 508 NLELIRSAKVFHYGSISLIVEPCRSAHLKAMEVAKDAGCLLSYDPNL 648
>TC10920 similar to UP|O04897 (O04897) Fructokinase , partial (35%)
Length = 596
Score = 164 bits (414), Expect = 3e-41
Identities = 82/130 (63%), Positives = 100/130 (76%)
Frame = +2
Query: 212 DVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYTKDFKGWVYGF 271
D+IK+S EEI LT+G DPYDD ++ KLFH NLKLLLVTEG +GCRYYTK+F G V G
Sbjct: 2 DIIKISEEEISFLTKGEDPYDDAVV-HKLFHPNLKLLLVTEGAEGCRYYTKEFSGRVMGM 178
Query: 272 EVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVTGRGAIPSLP 331
+V+A+DTTGAGDSFV G LS L+ + + E+ LR+AL FAN CGA TVT RGAIP+LP
Sbjct: 179 KVDAVDTTGAGDSFVAGVLSQLAVDLSLIQKEEQLRDALKFANVCGALTVTERGAIPALP 358
Query: 332 TKSSVLRVML 341
T+ +VL ML
Sbjct: 359 TRETVLNAML 388
>TC11135 homologue to UP|SCRK_SOLTU (P37829) Fructokinase , partial (33%)
Length = 402
Score = 123 bits (308), Expect = 5e-29
Identities = 61/105 (58%), Positives = 82/105 (78%)
Frame = +1
Query: 23 LVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGNDEF 82
L+V FGEM+I+ VPT+ GVSL++A + K+P GA A V++A++RLGG +AF+GK+G+DEF
Sbjct: 94 LIVSFGEMLIDFVPTVSGVSLAEAPGFLKAPGGAPANVAIAVTRLGGKAAFVGKLGDDEF 273
Query: 83 GHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMF 127
GHML+ ILK+N V G+ FD ARTALAF +LK DG+ EFMF
Sbjct: 274 GHMLAGILKENDVVADGISFDHGARTALAFVTLK--ADGEREFMF 402
>TC15313 similar to UP|Q9FLH8 (Q9FLH8) Fructokinase 1, partial (23%)
Length = 610
Score = 107 bits (266), Expect = 4e-24
Identities = 51/85 (60%), Positives = 67/85 (78%)
Frame = +1
Query: 257 CRYYTKDFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANAC 316
CRYYTK+F+G V G +V+ +DTTGAGD+FV G L +++ I++DEK LR+AL FAN C
Sbjct: 1 CRYYTKEFRGRVAGVKVKPVDTTGAGDAFVSGILYSIASDLSIFQDEKRLRKALYFANVC 180
Query: 317 GAATVTGRGAIPSLPTKSSVLRVML 341
GA TVT RGAIP+LPTK +VL+ +L
Sbjct: 181 GAITVTERGAIPALPTKEAVLQFLL 255
>BP068922
Length = 433
Score = 63.2 bits (152), Expect = 6e-11
Identities = 31/61 (50%), Positives = 42/61 (68%)
Frame = -3
Query: 280 GAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVTGRGAIPSLPTKSSVLRV 339
GAGDSFVG LS + I +DE LRE L ++NACGA+T T +GA P+LP + VL++
Sbjct: 431 GAGDSFVGSLLSSFVDDQSIPEDEPRLREVLKYSNACGASTTTEKGASPALPQEEDVLKL 252
Query: 340 M 340
+
Sbjct: 251 I 249
>TC17524 similar to UP|O04897 (O04897) Fructokinase , partial (12%)
Length = 581
Score = 58.9 bits (141), Expect = 1e-09
Identities = 26/39 (66%), Positives = 33/39 (83%)
Frame = +2
Query: 20 KGPLVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATA 58
+ PLVVCFGEM+I+ VPTI G+SL+DA A+KK+P GA A
Sbjct: 461 ESPLVVCFGEMLIDFVPTISGLSLADAPAFKKAPGGAPA 577
>TC18867 similar to UP|Q9ASV2 (Q9ASV2) At1g06730/F4H5_22, partial (25%)
Length = 748
Score = 45.8 bits (107), Expect = 1e-05
Identities = 35/104 (33%), Positives = 53/104 (50%), Gaps = 11/104 (10%)
Frame = +1
Query: 12 SKHGRKICKGPLVVCFGEMMINLVPTIDGVSLSDAEAYKK----SP-------AGATAIV 60
+KH G L V ++++N VP + SLS+ +AY + SP AG +
Sbjct: 313 AKHADVATLGNLCV---DIVLN-VPQLPPPSLSERKAYMERLASSPPPKKYWEAGGNCNM 480
Query: 61 SVAISRLGGSSAFIGKVGNDEFGHMLSDILKQNGVDNSGLLFDE 104
++A SRLG IG VGN+ +G LSD+L G+ G+ D+
Sbjct: 481 AIAASRLGLDCISIGHVGNEIYGKFLSDVLHDEGIGMVGMSTDD 612
>BP056137
Length = 472
Score = 43.5 bits (101), Expect = 5e-05
Identities = 22/36 (61%), Positives = 25/36 (69%)
Frame = -1
Query: 302 DEKILREALDFANACGAATVTGRGAIPSLPTKSSVL 337
DE LRE L FANACGA T T AIP+LPT++ L
Sbjct: 379 DEPRLREVLKFANACGAITTTK*CAIPALPTQADAL 272
>TC9644 homologue to UP|Q9M394 (Q9M394) Fructokinase-like protein, partial
(17%)
Length = 552
Score = 37.0 bits (84), Expect = 0.005
Identities = 19/59 (32%), Positives = 31/59 (52%)
Frame = +3
Query: 277 DTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVTGRGAIPSLPTKSS 335
D TG+GD+ V L L+ +++++ +L L FA A G GA+ PT+S+
Sbjct: 30 DRTGSGDAVVAAILRKLTTCPEMFENQDVLERQLRFAVAAGIIAQWTIGAVRGFPTESA 206
>TC7970 similar to UP|ADK2_ARATH (Q9LZG0) Adenosine kinase 2 (AK 2)
(Adenosine 5'-phosphotransferase 2) , partial (97%)
Length = 1482
Score = 35.8 bits (81), Expect = 0.011
Identities = 57/251 (22%), Positives = 104/251 (40%), Gaps = 20/251 (7%)
Frame = +2
Query: 69 GSSAFIGKVGNDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFY 128
G+++++G +G D++G ++ K GV N DE+ T + E
Sbjct: 305 GATSYMGCIGKDKYGEEMTKNSKLAGV-NVHYYEDENTPTGTCAVCVVGG-----ERSLI 466
Query: 129 RNPSADILFRSEEIDK----SLIKKATIFHYGSVSLIKEPSR----STHIEAINYAKMCG 180
N +A ++SE + K +L++KA F+ L P + H A N
Sbjct: 467 ANLAAANCYKSEHLKKPENWALVEKAKYFYIAGFFLTVSPESIQLVAEHAAANNKI---- 634
Query: 181 SILSYAPNLTVPLWPSTEAAREGIMSIWNYADVIKVSVEEIRILTEGNDPYDD---KMIM 237
+ NL+ P E ++ Y D + + E R ++ + D ++ +
Sbjct: 635 ----FTMNLSAPF--ICEFFKDAQEKALPYMDFVFGNETEARTFSKVHGWETDNVEEIAL 796
Query: 238 K-----KLFHHNLKLLLVTEGIKGCRYYTKDFKGWVYGF----EVEAIDTTGAGDSFVGG 288
K K + ++ ++T+G +D K ++ + + +DT GAGD+FVGG
Sbjct: 797 KISQWPKASGTHKRITVITQGADPV-CVAEDGKVTLFPVILLPKEKLVDTNGAGDAFVGG 973
Query: 289 FLSILSAHKHI 299
FL+ L K I
Sbjct: 974 FLAQLVREKPI 1006
>AV770215
Length = 351
Score = 30.8 bits (68), Expect(2) = 0.030
Identities = 14/20 (70%), Positives = 15/20 (75%)
Frame = -3
Query: 301 KDEKILREALDFANACGAAT 320
+DE LRE L FANACGA T
Sbjct: 286 EDEPRLREVLKFANACGAIT 227
Score = 22.3 bits (46), Expect(2) = 0.030
Identities = 9/11 (81%), Positives = 9/11 (81%)
Frame = -1
Query: 280 GAGDSFVGGFL 290
GAGDSFVG L
Sbjct: 351 GAGDSFVGALL 319
>TC11390 similar to UP|Q8L993 (Q8L993) Carbohydrate kinase-like protein,
partial (26%)
Length = 577
Score = 34.3 bits (77), Expect = 0.031
Identities = 23/84 (27%), Positives = 39/84 (46%)
Frame = +3
Query: 207 IWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYTKDFKG 266
+ NYAD+I + +E R L D ++ + + + L+ VT+G KG
Sbjct: 15 VGNYADLIFANSDEARALCNF-DAKENTASVTRYLSQFVPLVSVTDGPKGSYIGVNGEAV 191
Query: 267 WVYGFEVEAIDTTGAGDSFVGGFL 290
++ +DT GAGD++ G L
Sbjct: 192 YIPPSPCVPVDTCGAGDAYASGIL 263
>BP067421
Length = 409
Score = 33.5 bits (75), Expect = 0.053
Identities = 24/58 (41%), Positives = 28/58 (47%)
Frame = -2
Query: 274 EAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVTGRGAIPSLP 331
E ID TGAGD+F G L + AH + EK+ L FA AA GA LP
Sbjct: 348 ELIDPTGAGDAFAGAILYAICAH---FTREKM----LCFAANVAAAQCRALGARTGLP 196
>AV411163
Length = 414
Score = 32.3 bits (72), Expect = 0.12
Identities = 24/78 (30%), Positives = 37/78 (46%), Gaps = 2/78 (2%)
Frame = +1
Query: 211 ADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNL--KLLLVTEGIKGCRYYTKDFKGWV 268
+DV+ ++ +E LT DP + ++L + K ++V G +G T
Sbjct: 175 SDVLLLTSDEAESLTGIGDPI---LAGQELLKRGIRTKWVIVKMGSRGSILITTSRIACA 345
Query: 269 YGFEVEAIDTTGAGDSFV 286
F+V IDT G GDSFV
Sbjct: 346 PAFKVNVIDTVGCGDSFV 399
>TC8904 homologue to UP|G3PB_PEA (P12859) Glyceraldehyde 3-phosphate
dehydrogenase B, chloroplast precursor (NADP-dependent
glyceraldehydephosphate dehydrogenase subunit B) ,
partial (69%)
Length = 978
Score = 31.2 bits (69), Expect = 0.26
Identities = 18/71 (25%), Positives = 36/71 (50%)
Frame = +3
Query: 20 KGPLVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGN 79
+ PL + + E+ I++V GV + A K AGA ++ A ++ ++ V
Sbjct: 534 RDPLKLPWAELGIDIVIEGTGVFVDGPGAGKHIQAGAKKVIITAPAKGADIPTYVVGVNE 713
Query: 80 DEFGHMLSDIL 90
++GH ++DI+
Sbjct: 714 GDYGHNVADII 746
>TC13089
Length = 520
Score = 31.2 bits (69), Expect = 0.26
Identities = 15/37 (40%), Positives = 23/37 (61%)
Frame = +2
Query: 304 KILREALDFANACGAATVTGRGAIPSLPTKSSVLRVM 340
K +E L FA A + V +GA+PS+P + SVL ++
Sbjct: 41 KSXKECLRFAAAAASLCVQVKGAMPSMPDRKSVLDLL 151
>AV777933
Length = 339
Score = 30.0 bits (66), Expect = 0.59
Identities = 14/41 (34%), Positives = 24/41 (58%)
Frame = +2
Query: 123 PEFMFYRNPSADILFRSEEIDKSLIKKATIFHYGSVSLIKE 163
P +FY ++FR+ ++ KSL + T+F+ S SL K+
Sbjct: 83 PLHLFYSLDETLLIFRTTQMKKSLTLRGTVFNKSSHSLFKK 205
>AV422745
Length = 440
Score = 29.6 bits (65), Expect = 0.77
Identities = 21/99 (21%), Positives = 43/99 (43%), Gaps = 4/99 (4%)
Frame = +1
Query: 23 LVVCFGEMMINLVPTIDGVSLSD----AEAYKKSPAGATAIVSVAISRLGGSSAFIGKVG 78
+VV G + ++ + T+ D + + K G ++RLG + I K+
Sbjct: 16 IVVGCGGVAVDFLATVAAYPKPDDKIRSTSLKVQGGGNAGNALTCLARLGLNPRVITKIA 195
Query: 79 NDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKN 117
+D G + D L+ +GVD S ++ + + + + N
Sbjct: 196 DDSQGRGILDELQADGVDTSFIVVSKEGTSPFTYIIVDN 312
>TC11208 weakly similar to GB|AAM78055.1|21928053|AY125545 At4g29227
{Arabidopsis thaliana;}, partial (23%)
Length = 599
Score = 29.6 bits (65), Expect = 0.77
Identities = 16/42 (38%), Positives = 24/42 (57%)
Frame = +1
Query: 274 EAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANA 315
E IDTTGAGD+F+G + + + + E +L A + A A
Sbjct: 115 ELIDTTGAGDAFIGAVIYAICSK---FSPETMLSFAANVAGA 231
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,736,966
Number of Sequences: 28460
Number of extensions: 72511
Number of successful extensions: 335
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 329
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 331
length of query: 342
length of database: 4,897,600
effective HSP length: 91
effective length of query: 251
effective length of database: 2,307,740
effective search space: 579242740
effective search space used: 579242740
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC147014.14


