

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.6 + phase: 0
(663 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
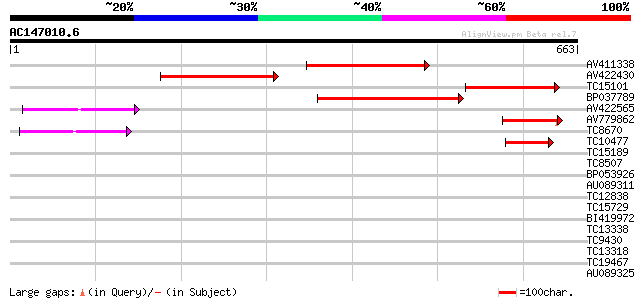
Score E
Sequences producing significant alignments: (bits) Value
AV411338 268 2e-72
AV422430 234 3e-62
TC15101 similar to UP|Q9LUS2 (Q9LUS2) Chloroplast outer envelope... 187 6e-48
BP037789 184 3e-47
AV422565 97 1e-20
AV779862 64 8e-11
TC8670 weakly similar to UP|Q9ZRD1 (Q9ZRD1) NTGP4 (Fragment), pa... 45 3e-05
TC10477 weakly similar to GB|AAA53276.1|576509|PEAIAP86A GTP-bin... 45 3e-05
TC15189 similar to UP|TO34_PEA (Q41009) Translocase of chloropla... 35 0.050
TC8507 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetyla... 32 0.25
BP053926 31 0.73
AU089311 31 0.73
TC12838 UP|Q40196 (Q40196) RAB11F, complete 30 0.95
TC15729 homologue to UP|RB1C_ARATH (O04486) Ras-related protein ... 30 1.2
BI419972 29 2.1
TC13338 similar to UP|Q9ZTZ4 (Q9ZTZ4) IFA-binding protein, parti... 29 2.8
TC9430 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase (L... 29 2.8
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 29 2.8
TC19467 homologue to UP|O82134 (O82134) Proliferating cell nucle... 29 2.8
AU089325 29 2.8
>AV411338
Length = 429
Score = 268 bits (685), Expect = 2e-72
Identities = 126/143 (88%), Positives = 139/143 (97%)
Frame = +1
Query: 348 KEMAESVKDLPSDYVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWL 407
K+MAES KD PSDY ENVEEESGGAASVPVPMPD++LPASFDSD PTHRYR+LDSSNQWL
Sbjct: 1 KKMAESAKDQPSDYSENVEEESGGAASVPVPMPDLALPASFDSDNPTHRYRYLDSSNQWL 180
Query: 408 VRPVLETHGWDHDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGE 467
VRPVLETHGWDHDVGYEGLNVERLFV+++K+P+SFSGQ+TKDKKDANVQME+TSS+K+GE
Sbjct: 181 VRPVLETHGWDHDVGYEGLNVERLFVVREKVPLSFSGQITKDKKDANVQMEVTSSIKHGE 360
Query: 468 GKATSLGFDMQTVGKDLAYTLRS 490
GKATSLGFDMQTVGKDLAYTLRS
Sbjct: 361 GKATSLGFDMQTVGKDLAYTLRS 429
>AV422430
Length = 415
Score = 234 bits (598), Expect = 3e-62
Identities = 117/138 (84%), Positives = 125/138 (89%)
Frame = +2
Query: 177 AAGDMRLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQ 236
AAGDMRLMNPVSLVENHSACRTN AGQRVLPNGQVWKP LLLLSFASKILAEANALLKLQ
Sbjct: 2 AAGDMRLMNPVSLVENHSACRTNRAGQRVLPNGQVWKPHLLLLSFASKILAEANALLKLQ 181
Query: 237 DNPREKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDP 296
D+P KPYT R+RAPPLPFLLS+LLQSRPQLKLPE+QF DEDS +DDLDE SDS DET+
Sbjct: 182 DSPPGKPYTTRSRAPPLPFLLSTLLQSRPQLKLPEEQFGDEDSPDDDLDESSDSDDETEL 361
Query: 297 DDLPPFKPLTKAQIRNLS 314
D+LPPFK LTKAQ+ LS
Sbjct: 362 DELPPFKALTKAQVEKLS 415
>TC15101 similar to UP|Q9LUS2 (Q9LUS2) Chloroplast outer envelope
protein-like, partial (10%)
Length = 616
Score = 187 bits (474), Expect = 6e-48
Identities = 90/109 (82%), Positives = 103/109 (93%)
Frame = +3
Query: 534 VIAGGAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPI 593
V++GGAM GR DVAYGGSLEAQLRDK+YPLGRSLSTLGLSVMDWHGDLAVGCN+QSQIP+
Sbjct: 3 VVSGGAMAGRGDVAYGGSLEAQLRDKDYPLGRSLSTLGLSVMDWHGDLAVGCNVQSQIPV 182
Query: 594 GRYTNLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVIGYSQ 642
GR+TNL+ARA+LNNRGAG ISIRLNSSEQLQIAL+GL PL++K+I +Q
Sbjct: 183 GRHTNLIARAHLNNRGAGPISIRLNSSEQLQIALVGLYPLIRKLISDAQ 329
>BP037789
Length = 548
Score = 184 bits (468), Expect = 3e-47
Identities = 84/170 (49%), Positives = 120/170 (70%)
Frame = +1
Query: 361 YVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGWDHD 420
Y E + E+G A+VPVP+PDM LP SFDS+ P +RYR L+ ++Q L RPVL++H WDHD
Sbjct: 31 YPEEDDPENGSPAAVPVPLPDMVLPPSFDSENPAYRYRFLEPTSQLLTRPVLDSHSWDHD 210
Query: 421 VGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQTV 480
GY+G+N+E+ + +K P + + QVTKDKKD ++ ++ + + K+G+ +T GFD+Q +
Sbjct: 211 CGYDGVNIEQTLAIINKFPGAVTVQVTKDKKDFSLHLDSSIAAKHGDNASTMAGFDIQNI 390
Query: 481 GKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKR 530
GK LAY +R ETKF NF RNK AG S T LG+ +S G+K ED++ KR
Sbjct: 391 GKQLAYIVRGETKFKNFKRNKTGAGFSVTFLGEDVSTGLKAEDQIALGKR 540
>AV422565
Length = 470
Score = 96.7 bits (239), Expect = 1e-20
Identities = 57/136 (41%), Positives = 75/136 (54%)
Frame = +3
Query: 16 SCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGIKVRVIDTPGLL 75
S TI+V+GK GVGKSST+NSI E + F + V G + +IDTPGL+
Sbjct: 69 SLTILVMGKGGVGKSSTVNSIIGERVVSISPFQSEGPRPVMVSRSRAGFTLNIIDTPGLI 248
Query: 76 PSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIFGPPIWF 135
N+ L+ +K F+ D++LY+DRLD D D + + ITD FG IW
Sbjct: 249 EGGYI---NDMALNLIKCFLLNKTIDVLLYVDRLDAYRVDNLDKLVAKAITDSFGKAIWN 419
Query: 136 NAIVVLTHAASAPPDG 151
AIV LTHA +PPDG
Sbjct: 420 RAIVALTHAQFSPPDG 467
>AV779862
Length = 534
Score = 63.9 bits (154), Expect = 8e-11
Identities = 33/75 (44%), Positives = 49/75 (65%), Gaps = 5/75 (6%)
Frame = -3
Query: 577 WHGDLAVGCNLQSQIPIGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKK 636
W GDLA+G +LQSQ +GR + RA LN + +GQI + +SSEQ+QIAL+ ++P+ +
Sbjct: 529 WRGDLALGADLQSQSSLGRSYKVAVRAGLNYKCSGQIRVSTSSSEQVQIALLAILPVARA 350
Query: 637 VI-----GYSQNCSL 646
V G S+N S+
Sbjct: 349 VYINFWPGASENYSI 305
>TC8670 weakly similar to UP|Q9ZRD1 (Q9ZRD1) NTGP4 (Fragment), partial
(71%)
Length = 1252
Score = 45.4 bits (106), Expect = 3e-05
Identities = 41/134 (30%), Positives = 63/134 (46%), Gaps = 3/134 (2%)
Frame = +2
Query: 12 PLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMG--TKKVQDVVGMVQGIKVRVI 69
P + T++++G++G GKS+T NSI + F +D G ++ + + G V VI
Sbjct: 83 PNNEGLTLVLVGRTGNGKSATANSIIGKKVFKSDYSSSGVTSRCERHTTELKDGQTVNVI 262
Query: 70 DTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSR-DFSDMPLLRTITDI 128
DTPGL + +E I + IK I L +QSR + LR++ +
Sbjct: 263 DTPGL---FDISAESEFIGKEIVNCIKLAKDGIHAVLVVFSVQSRFSKEEETALRSLQML 433
Query: 129 FGPPIWFNAIVVLT 142
FG I IVV T
Sbjct: 434 FGEKIADYMIVVFT 475
>TC10477 weakly similar to GB|AAA53276.1|576509|PEAIAP86A GTP-binding
protein {Pisum sativum;} , partial (5%)
Length = 625
Score = 45.4 bits (106), Expect = 3e-05
Identities = 21/56 (37%), Positives = 37/56 (65%)
Frame = +3
Query: 580 DLAVGCNLQSQIPIGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLK 635
++ + +LQS+ + R A+LN+R GQIS++++SSE LQIAL+G+ + +
Sbjct: 9 EMVLSGSLQSEFRLSRSLRASVGADLNSRKMGQISVKMSSSEHLQIALVGVFSIFQ 176
>TC15189 similar to UP|TO34_PEA (Q41009) Translocase of chloroplast 34 (34
kDa chloroplast outer envelope protein) (GTP-binding
protein OEP34) (GTP-binding protein IAP34), partial
(36%)
Length = 738
Score = 34.7 bits (78), Expect = 0.050
Identities = 14/32 (43%), Positives = 19/32 (58%)
Frame = +3
Query: 186 PVSLVENHSACRTNTAGQRVLPNGQVWKPQLL 217
PV L+EN C N + ++VLPNG W L+
Sbjct: 36 PVVLIENSGRCNKNDSDEKVLPNGIAWISNLV 131
>TC8507 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (40%)
Length = 620
Score = 32.3 bits (72), Expect = 0.25
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 4/42 (9%)
Frame = +3
Query: 271 EDQFSDEDSLNDDLDEPSD----SGDETDPDDLPPFKPLTKA 308
ED DED +DD DE D DE+D DD P+ KA
Sbjct: 9 EDDSDDEDDEDDDSDEEMDDADSDSDESDSDDTDEETPVKKA 134
>BP053926
Length = 562
Score = 30.8 bits (68), Expect = 0.73
Identities = 21/69 (30%), Positives = 34/69 (48%)
Frame = +1
Query: 18 TIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGIKVRVIDTPGLLPS 77
T+++ G VGKSS IN I + TK + + ++ +VIDTPG+L
Sbjct: 208 TVLICGYPNVGKSSFINKI-TRADVEVQPYAFTTKSLFVGHTDYKYLRYQVIDTPGIL-- 378
Query: 78 WSDQPHNEK 86
D+P ++
Sbjct: 379 --DRPFEDR 399
>AU089311
Length = 344
Score = 30.8 bits (68), Expect = 0.73
Identities = 23/79 (29%), Positives = 37/79 (46%), Gaps = 6/79 (7%)
Frame = -3
Query: 129 FGPPI--WFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRS-HVVQQAIR---QAAGDMR 182
F PP WF I++L + PP G + T S + + S +V++ + ++ D+
Sbjct: 309 FPPPSISWFGTIILLPLRSPEPPSGSSPTVSVFLALILYISVRIVKKHVSTLVDSSADVS 130
Query: 183 LMNPVSLVENHSACRTNTA 201
+ N SL N SA TA
Sbjct: 129 IKNKFSL*ANSSASSVGTA 73
>TC12838 UP|Q40196 (Q40196) RAB11F, complete
Length = 1099
Score = 30.4 bits (67), Expect = 0.95
Identities = 20/60 (33%), Positives = 30/60 (49%)
Frame = +2
Query: 8 AGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGIKVR 67
A E D+ +++G SGVGKS+ I+ +F D F + +K V + IKVR
Sbjct: 122 AFDEQCDYLFKAVLIGDSGVGKSNLIS------RFAKDEFRLDSKPTIGVEFAYRNIKVR 283
>TC15729 homologue to UP|RB1C_ARATH (O04486) Ras-related protein Rab11C,
partial (85%)
Length = 860
Score = 30.0 bits (66), Expect = 1.2
Identities = 20/80 (25%), Positives = 41/80 (51%), Gaps = 9/80 (11%)
Frame = +2
Query: 3 EQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTK---------K 53
E++ G+E D+ ++++G SGVGKS+ ++ +F + F + +K +
Sbjct: 77 EEMARRGEEEYDYLFKVVLIGDSGVGKSNLLS------RFTRNEFCLESKSTIGVEFATR 238
Query: 54 VQDVVGMVQGIKVRVIDTPG 73
+V G + +K ++ DT G
Sbjct: 239 TLEVEGRM--VKAQIWDTAG 292
>BI419972
Length = 498
Score = 29.3 bits (64), Expect = 2.1
Identities = 12/30 (40%), Positives = 18/30 (60%)
Frame = +1
Query: 271 EDQFSDEDSLNDDLDEPSDSGDETDPDDLP 300
ED +D+D ++ D D G+E DP+D P
Sbjct: 379 EDDDADDDDDDEGEDYSGDEGEEADPEDDP 468
>TC13338 similar to UP|Q9ZTZ4 (Q9ZTZ4) IFA-binding protein, partial (12%)
Length = 530
Score = 28.9 bits (63), Expect = 2.8
Identities = 21/82 (25%), Positives = 36/82 (43%)
Frame = +1
Query: 324 EVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSDYVENVEEESGGAASVPVPMPDMS 383
EVE +++ K+ +YE +++ MM S + S S + S ++S
Sbjct: 16 EVEKELEVYRKRVHEYEVREKMMMSRRDGSNNSMRS-------RTSSPSCSNAEDSDELS 174
Query: 384 LPASFDSDTPTHRYRHLDSSNQ 405
L + +S Y H +SSNQ
Sbjct: 175 LDLNHESKEENGSYSHQESSNQ 240
>TC9430 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase (LDH) ,
partial (49%)
Length = 866
Score = 28.9 bits (63), Expect = 2.8
Identities = 19/42 (45%), Positives = 23/42 (54%), Gaps = 2/42 (4%)
Frame = +3
Query: 144 AASAPPDGPNGTPSSYDMFVTQRSH--VVQQAIRQAAGDMRL 183
AA+ PP P+ TP S D VT S +V RQ AG+ RL
Sbjct: 483 AAALPPSHPDSTP-SVDYSVTAGSDLCIVTAGARQIAGESRL 605
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 28.9 bits (63), Expect = 2.8
Identities = 11/28 (39%), Positives = 18/28 (64%)
Frame = +3
Query: 271 EDQFSDEDSLNDDLDEPSDSGDETDPDD 298
+++ DED +DD D+ D G+E + DD
Sbjct: 270 DEEEDDEDEDDDDDDDDDDDGEEEEDDD 353
>TC19467 homologue to UP|O82134 (O82134) Proliferating cell nuclear antigen
(PCNA), partial (46%)
Length = 450
Score = 28.9 bits (63), Expect = 2.8
Identities = 21/85 (24%), Positives = 37/85 (42%)
Frame = +3
Query: 335 KQLKYEKKQRKMMKEMAESVKDLPSDYVENVEEESGGAASVPVPMPDMSLPASFDSDTPT 394
K L+ Q ++K++ ES+K+L +D A+FD T
Sbjct: 78 KMLELRLVQGSLLKKVLESIKELVND-------------------------ANFDCSTTG 182
Query: 395 HRYRHLDSSNQWLVRPVLETHGWDH 419
+ +DSS+ LV +L + G++H
Sbjct: 183 FSLQAMDSSHVALVALLLRSEGFEH 257
>AU089325
Length = 757
Score = 28.9 bits (63), Expect = 2.8
Identities = 20/69 (28%), Positives = 35/69 (49%), Gaps = 2/69 (2%)
Frame = +3
Query: 7 SAGQEP-LDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTK-KVQDVVGMVQGI 64
SA P D+ ++++G SGVGKS + D+ + +G K++ V + I
Sbjct: 39 SAAMNPEYDYLFKLLLIGDSGVGKSCLLLRFADDSYIESYISTIGVDFKIRTVEQDAKTI 218
Query: 65 KVRVIDTPG 73
K+++ DT G
Sbjct: 219 KLQIWDTAG 245
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,074,956
Number of Sequences: 28460
Number of extensions: 129322
Number of successful extensions: 807
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 770
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 796
length of query: 663
length of database: 4,897,600
effective HSP length: 96
effective length of query: 567
effective length of database: 2,165,440
effective search space: 1227804480
effective search space used: 1227804480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC147010.6


