

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.3 - phase: 0 /pseudo
(675 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
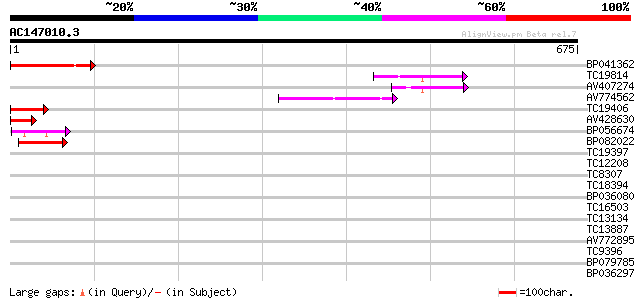
Score E
Sequences producing significant alignments: (bits) Value
BP041362 98 5e-21
TC19814 homologue to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, pa... 89 3e-18
AV407274 67 7e-12
AV774562 64 1e-10
TC19406 similar to UP|Q9FJS2 (Q9FJS2) Homeobox protein, partial ... 59 3e-09
AV428630 48 6e-06
BP056674 44 7e-05
BP082022 41 7e-04
TC19397 similar to PIR|S47137|S47137 homeotic protein Athb-7 - A... 39 0.003
TC12208 similar to UP|Q8LC03 (Q8LC03) Homeobox gene 13 protein, ... 39 0.003
TC8307 weakly similar to UP|Q9SP47 (Q9SP47) Homeodomain-leucine ... 39 0.004
TC18394 similar to UP|O23208 (O23208) Homeodomain protein, parti... 37 0.014
BP036080 34 0.088
TC16503 homologue to UP|HKL3_MALDO (O04136) Homeobox protein kno... 33 0.20
TC13134 similar to UP|O80409 (O80409) Pharbitis kntted-like gene... 33 0.20
TC13887 homologue to UP|Q8VY42 (Q8VY42) Hirzina, partial (14%) 33 0.20
AV772895 32 0.44
TC9396 similar to UP|Q9ZRB9 (Q9ZRB9) Homeobox 1 protein (Fragmen... 32 0.44
BP079785 31 0.57
BP036297 31 0.57
>BP041362
Length = 560
Score = 97.8 bits (242), Expect = 5e-21
Identities = 46/101 (45%), Positives = 67/101 (65%)
Frame = +3
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ QR +L+ ++GLEP Q+K WFQNKR +K QHER N +LR EN+K+R +N+
Sbjct: 237 KECPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQMKTQHERHENTSLRTENEKLRADNM 416
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
+ +E L C +CGG P + ++ ENA+L++E
Sbjct: 417 RYREALSNASCPNCGG-PTAXGEMSFDEHHLRXENARLREE 536
>TC19814 homologue to UP|Q94C37 (Q94C37) At1g05230/YUP8H12_16, partial (17%)
Length = 389
Score = 88.6 bits (218), Expect = 3e-18
Identities = 47/115 (40%), Positives = 69/115 (59%), Gaps = 3/115 (2%)
Frame = +3
Query: 434 IGVIQTLEGRNSVIKLADRMVKMFCECLTMPGQVELNHLTLDSIGGVRVSIRATTD---D 490
+GVI +GR S++KLA+RMV FC ++ T+ G +R T D
Sbjct: 51 VGVITNQDGRKSMLKLAERMVISFCAGVS--ASTAHTWTTISGTGAETDDVRVMTRKSVD 224
Query: 491 DASQPNGTVVTAATTLWLPLPAQKVFEFLKDPTKRSQWNGLSCGNPMHEIAHISN 545
D +P G V++AAT+ WLP+P ++VFEFL+D RS+W+ LS G + E+AHI+N
Sbjct: 225 DPGRPPGIVLSAATSFWLPVPPKRVFEFLRDENSRSEWDILSNGGVVQEMAHIAN 389
>AV407274
Length = 285
Score = 67.4 bits (163), Expect = 7e-12
Identities = 37/97 (38%), Positives = 57/97 (58%), Gaps = 5/97 (5%)
Frame = +2
Query: 455 KMFCE--CLTMPGQVELNHLTLDSIGGVRVSIRATTD---DDASQPNGTVVTAATTLWLP 509
K FC C + + EL HL G + +R T DD +P+G V++AAT++W+P
Sbjct: 2 KNFCSGVCASSVHKWELLHL-----GNMGDDMRVMTRKNLDDPGEPHGIVLSAATSVWMP 166
Query: 510 LPAQKVFEFLKDPTKRSQWNGLSCGNPMHEIAHISNG 546
+ ++F+FL+D RS+W+ LS G PM E+ HI+ G
Sbjct: 167 VSRHRLFDFLRDEQLRSEWDILSNGGPMQEMVHIAKG 277
>AV774562
Length = 460
Score = 63.5 bits (153), Expect = 1e-10
Identities = 44/142 (30%), Positives = 66/142 (45%), Gaps = 1/142 (0%)
Frame = +3
Query: 321 REFNIIRYCKKVDPGVWVITDVSFDSSRP-NTAPLSRGWKHPSGCIIREMPHGGCLVTWV 379
RE ++RYCK++ W I DVS D + +T P S ++R +
Sbjct: 3 REVYLVRYCKQLSGEQWAIVDVSIDKVKTTSTLPSSNAENAHPVALLRTSQMAIAK*SG* 182
Query: 380 EHVEVEDKIHTHYVYRDLVGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQT 439
E + K H +YR +V +GA WI LQ CER + +P++++ GV
Sbjct: 183 STCECQ-KGAVHTMYRAIVNSGLAFGARHWIATLQLQCERLVFFMATNVPMKDSTGVATW 359
Query: 440 LEGRNSVIKLADRMVKMFCECL 461
E R S++KLA RM FC +
Sbjct: 360 AE-RKSILKLAQRMTWGFCHAV 422
>TC19406 similar to UP|Q9FJS2 (Q9FJS2) Homeobox protein, partial (10%)
Length = 546
Score = 58.5 bits (140), Expect = 3e-09
Identities = 23/45 (51%), Positives = 33/45 (73%)
Frame = +1
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETN 46
KEC HPD+ QR +L+ +GL+P+Q+K WFQN+R +K Q +R N
Sbjct: 403 KECPHPDDKQRMKLSHDLGLKPRQVKFWFQNRRTQMKAQQDRSDN 537
>AV428630
Length = 342
Score = 47.8 bits (112), Expect = 6e-06
Identities = 19/31 (61%), Positives = 25/31 (80%)
Frame = +1
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQN 32
KEC HPD+ QR +L+ ++GLEP Q+K WFQN
Sbjct: 250 KECPHPDDKQRKELSRELGLEPLQVKFWFQN 342
>BP056674
Length = 566
Score = 44.3 bits (103), Expect = 7e-05
Identities = 26/78 (33%), Positives = 39/78 (49%), Gaps = 8/78 (10%)
Frame = +1
Query: 3 ECHHPDEAQRCQLA----VKIGLEPKQIKSWFQNKRAMLKHQHE----RETNGTLRREND 54
EC P +R QL V +EPKQIK WFQN+R K + E + N L N
Sbjct: 172 ECPKPSSLRRQQLIRECPVLANVEPKQIKVWFQNRRCREKQRKEASRLQAVNRKLNAMNK 351
Query: 55 KIRNENLKIKEELKAKIC 72
+ EN ++++++ +C
Sbjct: 352 LLMEENDRLQKQVSQLVC 405
>BP082022
Length = 455
Score = 40.8 bits (94), Expect = 7e-04
Identities = 18/58 (31%), Positives = 37/58 (63%)
Frame = -3
Query: 11 QRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENLKIKEELK 68
Q+ LA ++ L P+Q++ WFQN+RA K + L++ + + +EN+++++EL+
Sbjct: 402 QKQALARQLNLRPRQVEVWFQNRRARTKLKQTEVDCDFLKKCCETLTDENMRLQKELQ 229
>TC19397 similar to PIR|S47137|S47137 homeotic protein Athb-7 - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (35%)
Length = 723
Score = 38.9 bits (89), Expect = 0.003
Identities = 19/35 (54%), Positives = 26/35 (74%), Gaps = 1/35 (2%)
Frame = +2
Query: 11 QRCQLAVKIGLEPKQIKSWFQNKRAMLK-HQHERE 44
++ QLA ++GL+P+Q+ WFQNKRA K Q ERE
Sbjct: 335 KKLQLARELGLQPRQVAIWFQNKRARWKSKQLERE 439
>TC12208 similar to UP|Q8LC03 (Q8LC03) Homeobox gene 13 protein, partial
(24%)
Length = 660
Score = 38.9 bits (89), Expect = 0.003
Identities = 16/31 (51%), Positives = 23/31 (73%)
Frame = +1
Query: 11 QRCQLAVKIGLEPKQIKSWFQNKRAMLKHQH 41
++ QLA +GL+P+QI WFQN+RA K +H
Sbjct: 562 RKMQLAKALGLQPRQIAIWFQNRRARWKTKH 654
>TC8307 weakly similar to UP|Q9SP47 (Q9SP47) Homeodomain-leucine zipper
protein 57, partial (51%)
Length = 1629
Score = 38.5 bits (88), Expect = 0.004
Identities = 19/64 (29%), Positives = 38/64 (58%), Gaps = 3/64 (4%)
Frame = +1
Query: 11 QRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNEN---LKIKEEL 67
++ ++A +GL+P+Q+ WFQN+RA K + + +L + +++ LK K+ L
Sbjct: 718 RKTKIAKDLGLQPRQVAIWFQNRRARWKTKQLEKDYDSLHSSFESLKSNYDNLLKEKDML 897
Query: 68 KAKI 71
KA++
Sbjct: 898 KAEV 909
>TC18394 similar to UP|O23208 (O23208) Homeodomain protein, partial (46%)
Length = 846
Score = 36.6 bits (83), Expect = 0.014
Identities = 15/47 (31%), Positives = 29/47 (60%)
Frame = +2
Query: 5 HHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRR 51
H + ++ +LA ++ L+P+Q+ WFQN+RA K++ E L++
Sbjct: 233 HKLESERKDKLAAELCLDPRQVAVWFQNRRARWKNKKLEEEYSNLKK 373
>BP036080
Length = 409
Score = 33.9 bits (76), Expect = 0.088
Identities = 14/29 (48%), Positives = 20/29 (68%)
Frame = -3
Query: 6 HPDEAQRCQLAVKIGLEPKQIKSWFQNKR 34
+P E+Q+ LA GL+ KQI +WF N+R
Sbjct: 377 YPSESQKLALAEATGLDQKQINNWFINQR 291
>TC16503 homologue to UP|HKL3_MALDO (O04136) Homeobox protein knotted-1 like
3 (KNAP3), partial (22%)
Length = 723
Score = 32.7 bits (73), Expect = 0.20
Identities = 20/66 (30%), Positives = 31/66 (46%), Gaps = 9/66 (13%)
Frame = +1
Query: 6 HPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDK---------I 56
+P E + +L + GL+ KQI +WF N+R ++ H + T + K I
Sbjct: 130 YPTEEDKARLVQETGLQLKQINNWFINQRK--RNWHSNASTSTALKSKRKRS**C*WLHI 303
Query: 57 RNENLK 62
ENLK
Sbjct: 304 NGENLK 321
>TC13134 similar to UP|O80409 (O80409) Pharbitis kntted-like gene 2, partial
(29%)
Length = 502
Score = 32.7 bits (73), Expect = 0.20
Identities = 13/29 (44%), Positives = 20/29 (68%)
Frame = +2
Query: 6 HPDEAQRCQLAVKIGLEPKQIKSWFQNKR 34
+P E+ + +LA GL+ KQI +WF N+R
Sbjct: 119 YPTESDKIELAKATGLDQKQINNWFINQR 205
>TC13887 homologue to UP|Q8VY42 (Q8VY42) Hirzina, partial (14%)
Length = 514
Score = 32.7 bits (73), Expect = 0.20
Identities = 14/29 (48%), Positives = 20/29 (68%)
Frame = +1
Query: 6 HPDEAQRCQLAVKIGLEPKQIKSWFQNKR 34
+P E+Q+ LA GL+ KQI +WF N+R
Sbjct: 1 YPSESQKQALAESTGLDLKQINNWFINQR 87
>AV772895
Length = 103
Score = 31.6 bits (70), Expect = 0.44
Identities = 11/22 (50%), Positives = 18/22 (81%)
Frame = -3
Query: 376 VTWVEHVEVEDKIHTHYVYRDL 397
+TW+EHVE+EDK + +YR++
Sbjct: 86 LTWLEHVEIEDKTPVNRLYRNV 21
>TC9396 similar to UP|Q9ZRB9 (Q9ZRB9) Homeobox 1 protein (Fragment),
partial (68%)
Length = 1138
Score = 31.6 bits (70), Expect = 0.44
Identities = 14/43 (32%), Positives = 22/43 (50%)
Frame = +1
Query: 6 HPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGT 48
+P E + +L + GL+ KQI +WF N+R H + T
Sbjct: 745 YPTEEDKARLVQETGLQLKQINNWFINQRKRNWHTNPSSATST 873
>BP079785
Length = 472
Score = 31.2 bits (69), Expect = 0.57
Identities = 13/29 (44%), Positives = 19/29 (64%)
Frame = -3
Query: 6 HPDEAQRCQLAVKIGLEPKQIKSWFQNKR 34
+P E ++ QL+ GL+ KQI WF N+R
Sbjct: 365 YPTEEEKLQLSEMTGLDIKQINHWFINQR 279
>BP036297
Length = 569
Score = 31.2 bits (69), Expect = 0.57
Identities = 15/49 (30%), Positives = 28/49 (56%)
Frame = +3
Query: 6 HPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRREND 54
+P E R +L+ K+GL +Q++ WF ++R + + T G+ + ND
Sbjct: 240 YPGEVLRAELSEKLGLSDRQLQMWFCHRRL----KDRKATPGSNKLLND 374
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.330 0.142 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,857,151
Number of Sequences: 28460
Number of extensions: 192585
Number of successful extensions: 1087
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 1080
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1084
length of query: 675
length of database: 4,897,600
effective HSP length: 96
effective length of query: 579
effective length of database: 2,165,440
effective search space: 1253789760
effective search space used: 1253789760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC147010.3


