

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146972.1 + phase: 0 /pseudo
(594 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
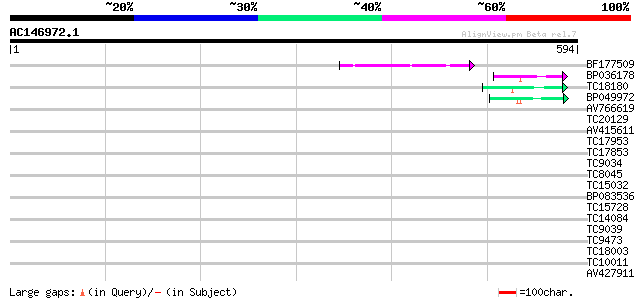
BF177509 50 1e-06
BP036178 42 2e-04
TC18180 weakly similar to GB|AAF04891.1|6175165|ATAC011437 Mutat... 41 6e-04
BP049972 40 8e-04
AV766619 40 0.001
TC20129 homologue to GB|AAF04891.1|6175165|ATAC011437 Mutator-li... 39 0.002
AV415611 36 0.015
TC17953 similar to GB|AAM26637.1|20855902|AY101516 AT3g06940/F17... 36 0.020
TC17853 similar to UP|Q42412 (Q42412) RNA-binding protein RZ-1, ... 35 0.045
TC9034 34 0.077
TC8045 weakly similar to UP|ST35_ARATH (Q94LW6) Probable sulfate... 30 0.85
TC15032 similar to UP|O81340 (O81340) 26S proteasome regulatory ... 30 1.1
BP083536 28 3.2
TC15728 similar to AAS09986 (AAS09986) MYB transcription factor,... 28 4.2
TC14084 28 4.2
TC9039 28 4.2
TC9473 similar to AAS09986 (AAS09986) MYB transcription factor, ... 28 5.5
TC18003 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 28 5.5
TC10011 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, par... 27 7.2
AV427911 27 7.2
>BF177509
Length = 472
Score = 50.1 bits (118), Expect = 1e-06
Identities = 41/142 (28%), Positives = 59/142 (40%)
Frame = +1
Query: 346 YLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIRCYVMRKMS 405
++ + P W AYF E + ++ N E N+ IL G PI+ M E IR +M +
Sbjct: 19 WIRRIPPRLWATAYF-EGQRFGHLTANIVESLNTWILEASGLPIIQMMECIRRQLMTWFN 195
Query: 406 QNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCRIKVDVDIFK 465
+ + A L P R E + + + A I VDI
Sbjct: 196 ERRETSMQWASILVP-SAERCVAEALERARTYQVLRANEAEFEVISHEGSNI---VDIRN 363
Query: 466 QTCTCRFWQLT*MPCMHACSAL 487
+ C CR WQL +PC HA +AL
Sbjct: 364 RCCLCRGWQLYGLPCAHAVAAL 429
>BP036178
Length = 561
Score = 42.4 bits (98), Expect = 2e-04
Identities = 28/89 (31%), Positives = 38/89 (42%), Gaps = 11/89 (12%)
Frame = -2
Query: 507 YRETYSYFIQPMNSHIYW-EPTPYEKPV----------PPKVKRSAGRPKKNRRKDGNEE 555
YR YS I P+ W E + V PPK +R GRPKK + N
Sbjct: 560 YRMAYSQIIIPIPDRSQWREHGEGAEGVGGARVDIVIRPPKTRRPPGRPKKKVLRVEN-- 387
Query: 556 PIGGSSMKKTYNDIQCGRCGLMGHNSRSC 584
K+ +QCGRC ++GH+ + C
Sbjct: 386 ------FKRPKRIVQCGRCHMLGHSQKKC 318
>TC18180 weakly similar to GB|AAF04891.1|6175165|ATAC011437 Mutator-like
transposase {Arabidopsis thaliana;} , partial (4%)
Length = 513
Score = 40.8 bits (94), Expect = 6e-04
Identities = 27/92 (29%), Positives = 36/92 (38%), Gaps = 3/92 (3%)
Frame = +3
Query: 496 DHCHAWLTLGSYRETYSYFIQPMNSHIYWE---PTPYEKPVPPKVKRSAGRPKKNRRKDG 552
D+C + T SYR TYS + P+ E P PP +R GRP R
Sbjct: 12 DYCSRYCTTESYRLTYSERVNPIPIMAVPESKDSQPVVTVTPPFTRRPPGRPATKRY--- 182
Query: 553 NEEPIGGSSMKKTYNDIQCGRCGLMGHNSRSC 584
S ++ C RC +GHN +C
Sbjct: 183 -------VSQNIVKRELHCSRCKGLGHNKSTC 257
>BP049972
Length = 496
Score = 40.4 bits (93), Expect = 8e-04
Identities = 28/102 (27%), Positives = 38/102 (36%), Gaps = 19/102 (18%)
Frame = -1
Query: 503 TLGSYRETYSYFIQPMNSHIYWEPTPYE-----KPV--------------PPKVKRSAGR 543
T+ +YR+TYS I P+ +W E K V PPK R GR
Sbjct: 472 TVATYRKTYSQTIHPIPDRSFWNELSIEDANANKAVEDANASQSIEIVINPPKSLRPPGR 293
Query: 544 PKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGLMGHNSRSCA 585
P+K R + + + C RC GH +CA
Sbjct: 292 PRKKR--------VRAEDRGRVKRVVHCSRCNQTGHFRTTCA 191
>AV766619
Length = 412
Score = 40.0 bits (92), Expect = 0.001
Identities = 30/99 (30%), Positives = 47/99 (47%), Gaps = 7/99 (7%)
Frame = +2
Query: 270 FVVYCLNWQGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNK---ELRRVV*E--CARET 324
FV C N GL ++ E+ H YC+ +L + K K + E RR + A
Sbjct: 26 FVANCQN--GLKKSLSEIFEKCHHSYCLRYLAEKLNKDLKGQFSHEARRFMINDFYAAAY 199
Query: 325 TPTQ--FNRIMERVKSINQKAWEYLNKWPKEAWIKAYFN 361
P Q F R +E +K I+ +A+ ++ + E W A+FN
Sbjct: 200 APKQETFERSVENMKGISSEAFNWVMQNEPEHWANAFFN 316
>TC20129 homologue to GB|AAF04891.1|6175165|ATAC011437 Mutator-like
transposase {Arabidopsis thaliana;} , partial (12%)
Length = 588
Score = 38.9 bits (89), Expect = 0.002
Identities = 27/93 (29%), Positives = 36/93 (38%), Gaps = 10/93 (10%)
Frame = +3
Query: 503 TLGSYRETYSYFIQPMNSHIYW-------EPTPYEKPV---PPKVKRSAGRPKKNRRKDG 552
T+ +YR+TYS I P+ W E V PPK R GRP+K R
Sbjct: 15 TVATYRKTYSQTIHPIPDKSLWKELSEGDENAAKADQVLINPPKSLRPPGRPRKKR---- 182
Query: 553 NEEPIGGSSMKKTYNDIQCGRCGLMGHNSRSCA 585
+ + + C RC GH +CA
Sbjct: 183 ----VRAEDRGRVKRVVHCSRCNQTGHFRTTCA 269
>AV415611
Length = 219
Score = 36.2 bits (82), Expect = 0.015
Identities = 21/65 (32%), Positives = 35/65 (53%)
Frame = +3
Query: 423 QSRLEKEKIASHNWTPLWNGDNARQRYHIENNCRIKVDVDIFKQTCTCRFWQLT*MPCMH 482
+ +L++E SH PL + Y + + +V VDI + C+C+ WQLT +PC H
Sbjct: 9 EEKLKRESQKSH---PLQVILSTGSTYEVCGDTPTEV-VDIDRWECSCKTWQLTGVPCCH 176
Query: 483 ACSAL 487
A + +
Sbjct: 177 AIAVI 191
>TC17953 similar to GB|AAM26637.1|20855902|AY101516 AT3g06940/F17A9_9
{Arabidopsis thaliana;}, partial (6%)
Length = 527
Score = 35.8 bits (81), Expect = 0.020
Identities = 18/51 (35%), Positives = 25/51 (48%)
Frame = +3
Query: 534 PPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGLMGHNSRSC 584
PP KR GRPK +P+ M K +QC +C +GHN ++C
Sbjct: 42 PPPTKRPPGRPKM--------KPVESIDMIK--RQLQCSKCKELGHNKKTC 164
>TC17853 similar to UP|Q42412 (Q42412) RNA-binding protein RZ-1, partial
(58%)
Length = 881
Score = 34.7 bits (78), Expect = 0.045
Identities = 19/56 (33%), Positives = 26/56 (45%)
Frame = +2
Query: 539 RSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGLMGHNSRSCAKQGVARRPK 594
R GR + +R GGS + N +C +CG GH +R C +G RR K
Sbjct: 332 RERGRDRDDRGDRDRSRGYGGS---RGSNGGECFKCGKPGHFARECPSEGETRRGK 490
>TC9034
Length = 583
Score = 33.9 bits (76), Expect = 0.077
Identities = 18/66 (27%), Positives = 31/66 (46%)
Frame = +1
Query: 529 YEKPVPPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGLMGHNSRSCAKQG 588
Y K P +KR G + NR++ E P ++++ ++C C GH+S C +
Sbjct: 25 YGKIDSPTLKRLPGWERINRKRGVTEGPTVSHEARRSHR-VKCANCEEFGHDSWECQRDK 201
Query: 589 VARRPK 594
R+ K
Sbjct: 202 SKRQEK 219
>TC8045 weakly similar to UP|ST35_ARATH (Q94LW6) Probable sulfate
transporter 3.5, partial (39%)
Length = 1616
Score = 30.4 bits (67), Expect = 0.85
Identities = 13/36 (36%), Positives = 19/36 (52%)
Frame = +2
Query: 467 TCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWL 502
TC + W++ *+ + C A+ F DHC AWL
Sbjct: 749 TCNMQAWKVK*VWYIQRC*AIPCFNFPRIDHCSAWL 856
>TC15032 similar to UP|O81340 (O81340) 26S proteasome regulatory subunit
S5A, partial (24%)
Length = 653
Score = 30.0 bits (66), Expect = 1.1
Identities = 17/50 (34%), Positives = 29/50 (58%)
Frame = -2
Query: 474 QLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQPMNSHIY 523
Q T +P +++C L GF L HCH LT+G + T + +Q + +++Y
Sbjct: 217 QTTDLPTIYSC----LTGF-LSPHCHLLLTIGELKLTQDH-LQLLQTYLY 86
>BP083536
Length = 377
Score = 28.5 bits (62), Expect = 3.2
Identities = 9/25 (36%), Positives = 14/25 (56%)
Frame = +1
Query: 466 QTCTCRFWQLT*MPCMHACSALALR 490
+ C C W+L +PC HA + + R
Sbjct: 301 KACCCNLWELVEIPCRHALAGVHYR 375
>TC15728 similar to AAS09986 (AAS09986) MYB transcription factor, partial
(34%)
Length = 1207
Score = 28.1 bits (61), Expect = 4.2
Identities = 10/20 (50%), Positives = 12/20 (60%)
Frame = +1
Query: 571 CGRCGLMGHNSRSCAKQGVA 590
C +CG GHNSR+C A
Sbjct: 199 CSQCGNNGHNSRTCTDTAAA 258
>TC14084
Length = 763
Score = 28.1 bits (61), Expect = 4.2
Identities = 14/42 (33%), Positives = 22/42 (52%), Gaps = 8/42 (19%)
Frame = +3
Query: 500 AWLTLGSYRETYSYFIQPMNS--------HIYWEPTPYEKPV 533
+WL L +YR+ Y I+ +N H+YW P+P P+
Sbjct: 294 SWLPLQNYRK-YHPSIEEVNEPLSVTTPHHVYWPPSPLLPPI 416
>TC9039
Length = 1218
Score = 28.1 bits (61), Expect = 4.2
Identities = 15/58 (25%), Positives = 22/58 (37%)
Frame = +2
Query: 537 VKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGLMGHNSRSCAKQGVARRPK 594
V S + K+ + + E + KK D C C + GH + C K R K
Sbjct: 743 VSTSKDKGKRKKTVEPKNEAADAPAPKKQKEDDTCYFCNVSGHMKKKCTKYHAWRARK 916
>TC9473 similar to AAS09986 (AAS09986) MYB transcription factor, partial
(50%)
Length = 887
Score = 27.7 bits (60), Expect = 5.5
Identities = 11/35 (31%), Positives = 20/35 (56%)
Frame = +3
Query: 555 EPIGGSSMKKTYNDIQCGRCGLMGHNSRSCAKQGV 589
+P+ ++M + +C C GHN+R+C +GV
Sbjct: 162 DPLRNAAMTR-----RCSHCSHGGHNARTCPNRGV 251
>TC18003 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (42%)
Length = 587
Score = 27.7 bits (60), Expect = 5.5
Identities = 10/24 (41%), Positives = 15/24 (61%)
Frame = +1
Query: 568 DIQCGRCGLMGHNSRSCAKQGVAR 591
D++C CG GH +R C +G +R
Sbjct: 157 DLKCYECGEPGHFARECRSRGGSR 228
>TC10011 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (62%)
Length = 684
Score = 27.3 bits (59), Expect = 7.2
Identities = 16/48 (33%), Positives = 21/48 (43%)
Frame = +2
Query: 537 VKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGLMGHNSRSC 584
V+ + G P+ NR G P G +C CGL GH +R C
Sbjct: 296 VEFAKGGPRGNREYLGRGPPPGSG---------RCFNCGLDGHWARDC 412
>AV427911
Length = 348
Score = 27.3 bits (59), Expect = 7.2
Identities = 11/29 (37%), Positives = 16/29 (54%)
Frame = +1
Query: 508 RETYSYFIQPMNSHIYWEPTPYEKPVPPK 536
++ Y Y P H+Y +PT + P PPK
Sbjct: 211 KKPYKYPSPPPPVHVYPKPTYHSPPPPPK 297
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.350 0.154 0.574
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,367,362
Number of Sequences: 28460
Number of extensions: 204491
Number of successful extensions: 2143
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 2098
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2133
length of query: 594
length of database: 4,897,600
effective HSP length: 95
effective length of query: 499
effective length of database: 2,193,900
effective search space: 1094756100
effective search space used: 1094756100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146972.1


