

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.10 - phase: 0
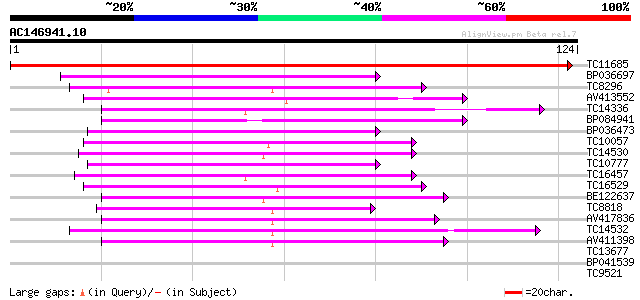
(124 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC11685 homologue to UP|PM14_ARATH (Q9FMP4) Pre-mRNA branch site... 243 5e-66
BP036697 48 6e-07
TC8296 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1... 47 1e-06
AV413552 47 1e-06
TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, p... 45 5e-06
BP084941 44 6e-06
BP036473 44 6e-06
TC10057 similar to UP|Q41124 (Q41124) Chloroplast RNA binding pr... 44 1e-05
TC14530 similar to UP|P82277 (P82277) Plastid-specific 30S ribos... 43 1e-05
TC10777 similar to UP|Q9SCL3 (Q9SCL3) Pre-mRNA splicing factor S... 43 2e-05
TC16457 similar to UP|Q41124 (Q41124) Chloroplast RNA binding pr... 42 2e-05
TC16529 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, p... 41 7e-05
BE122637 40 9e-05
TC8818 weakly similar to PIR|T48456|T48456 rna binding protein-l... 40 1e-04
AV417836 39 3e-04
TC14532 similar to UP|Q9FYA8 (Q9FYA8) SC35-like splicing factor ... 39 3e-04
AV411398 37 8e-04
TC13677 similar to UP|Q8H0P8 (Q8H0P8) RNA-binding protein, parti... 37 0.001
BP041539 35 0.003
TC9521 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, part... 35 0.004
>TC11685 homologue to UP|PM14_ARATH (Q9FMP4) Pre-mRNA branch site p14-like
protein, complete
Length = 558
Score = 243 bits (621), Expect = 5e-66
Identities = 121/123 (98%), Positives = 121/123 (98%)
Frame = +1
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M TISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGT KDTRGTA
Sbjct: 43 MATISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTTKDTRGTA 222
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS
Sbjct: 223 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 402
Query: 121 TKD 123
TKD
Sbjct: 403 TKD 411
>BP036697
Length = 578
Score = 47.8 bits (112), Expect = 6e-07
Identities = 25/70 (35%), Positives = 38/70 (53%)
Frame = +3
Query: 12 RLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAK 71
++ +R +YV NLP +I E+ D+F KYG I I + G AFV +ED DA+
Sbjct: 75 KMSRRTSRTVYVGNLPGDIREREVEDLFYKYGNITHIDLKVPPRPPGYAFVEFEDTQDAE 254
Query: 72 TAVDHLSGFN 81
A+ G++
Sbjct: 255 DAIRGRDGYD 284
>TC8296 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1_4
{Arabidopsis thaliana;}, partial (55%)
Length = 1276
Score = 46.6 bits (109), Expect = 1e-06
Identities = 29/83 (34%), Positives = 45/83 (53%), Gaps = 5/83 (6%)
Frame = +2
Query: 14 PPEVNRV--LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIY 68
PP++ L V N+ F T+++++ +F KYG + I I ++ T RG AFV Y+
Sbjct: 110 PPDITDTYSLLVLNITFRTTADDLFPLFDKYGKVVDIFIPKDRRTGESRGFAFVRYKYAD 289
Query: 69 DAKTAVDHLSGFNVANRYLIVLY 91
+A AVD L G V R + V +
Sbjct: 290 EASKAVDRLDGRMVDGREITVQF 358
>AV413552
Length = 285
Score = 46.6 bits (109), Expect = 1e-06
Identities = 28/86 (32%), Positives = 47/86 (54%), Gaps = 2/86 (2%)
Frame = +1
Query: 17 VNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGT--AFVVYEDIYDAKTAV 74
+ + LYV NLP+++++ ++ D+FG+ G + + I KD RG AFV +A+ AV
Sbjct: 13 IKKKLYVFNLPWSMSAADIKDLFGQCGTVTDVEIIRGKDGRGKGYAFVTMASGEEAQAAV 192
Query: 75 DHLSGFNVANRYLIVLYYQQAKMSKK 100
D ++ R +L + AK KK
Sbjct: 193 DKFDTLELSGR---ILRVELAKRFKK 261
>TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(85%)
Length = 1917
Score = 44.7 bits (104), Expect = 5e-06
Identities = 29/99 (29%), Positives = 50/99 (50%), Gaps = 2/99 (2%)
Frame = +2
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRI--GTNKDTRGTAFVVYEDIYDAKTAVDHLS 78
LYV+NL +I E++ ++F YG I ++ N +RG+ FV + +A A+ ++
Sbjct: 680 LYVKNLDDSIADEKLKELFSSYGTITSCKVMRDPNGISRGSGFVAFSTPEEASRALLEMN 859
Query: 79 GFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKY 117
G V ++ L V Q +KED AR+Q ++
Sbjct: 860 GKMVVSKPLYVTLAQ-----------RKEDRRARLQAQF 943
>BP084941
Length = 435
Score = 44.3 bits (103), Expect = 6e-06
Identities = 26/80 (32%), Positives = 45/80 (55%)
Frame = -3
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHLSGF 80
L+V NL ++T ++ D+F +YGA+ + T +R AFV ++ + DAK A + L GF
Sbjct: 235 LWVGNLAPDVTDSDLMDLFAQYGALDSVTCYT---SRSYAFVFFKRVDDAKAAKNALQGF 65
Query: 81 NVANRYLIVLYYQQAKMSKK 100
++ L + + AK K+
Sbjct: 64 SLRGVSLKIEFAFPAKPCKQ 5
>BP036473
Length = 510
Score = 44.3 bits (103), Expect = 6e-06
Identities = 24/64 (37%), Positives = 36/64 (55%)
Frame = +3
Query: 18 NRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHL 77
+R +YV NLP +I E+ DIF K+G I + G AFV +ED DA+ A+ +
Sbjct: 69 SRTIYVGNLPGDIRLREVEDIFYKFGPXVDIDLKIPPRPPGYAFVQFEDARDAEDAIYYR 248
Query: 78 SGFN 81
G++
Sbjct: 249 DGYD 260
>TC10057 similar to UP|Q41124 (Q41124) Chloroplast RNA binding protein
precursor, partial (49%)
Length = 598
Score = 43.5 bits (101), Expect = 1e-05
Identities = 24/76 (31%), Positives = 40/76 (52%), Gaps = 3/76 (3%)
Frame = +2
Query: 17 VNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD---TRGTAFVVYEDIYDAKTA 73
VN LY NLP+++ S ++ + +YG+ I + ++D +RG AFV + D T
Sbjct: 308 VNTKLYFGNLPYSVDSAQLAGLIEEYGSAELIEVLYDRDSGKSRGFAFVTMSCVEDCNTV 487
Query: 74 VDHLSGFNVANRYLIV 89
+++L G R L V
Sbjct: 488 IENLDGKEFLGRTLRV 535
>TC14530 similar to UP|P82277 (P82277) Plastid-specific 30S ribosomal
protein 2, chloroplast precursor (PSRP-2), partial (68%)
Length = 1104
Score = 43.1 bits (100), Expect = 1e-05
Identities = 23/77 (29%), Positives = 44/77 (56%), Gaps = 3/77 (3%)
Frame = +2
Query: 16 EVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNK---DTRGTAFVVYEDIYDAKT 72
E +R LYV N+P +T++E+ +I ++ A+ + + +K +R AFV + + DA
Sbjct: 221 EADRRLYVGNIPRTLTNDELANIVQEHAAVEKAEVMYDKYSGRSRRFAFVTVKTVEDANA 400
Query: 73 AVDHLSGFNVANRYLIV 89
A++ L+G + R + V
Sbjct: 401 AIEKLNGTQIGGREIKV 451
>TC10777 similar to UP|Q9SCL3 (Q9SCL3) Pre-mRNA splicing factor SF2-like
protein, partial (46%)
Length = 597
Score = 42.7 bits (99), Expect = 2e-05
Identities = 23/64 (35%), Positives = 34/64 (52%)
Frame = +1
Query: 18 NRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHL 77
+R +YV NLP +I E+ DIF KYG I I + FV +++ DA+ A+
Sbjct: 253 SRTIYVGNLPGDIRESEIEDIFYKYGRILDIELKVPPRPPCYCFVEFDNARDAEDAIRGR 432
Query: 78 SGFN 81
G+N
Sbjct: 433 DGYN 444
>TC16457 similar to UP|Q41124 (Q41124) Chloroplast RNA binding protein
precursor, partial (30%)
Length = 655
Score = 42.4 bits (98), Expect = 2e-05
Identities = 24/78 (30%), Positives = 39/78 (49%), Gaps = 3/78 (3%)
Frame = +2
Query: 15 PEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRI---GTNKDTRGTAFVVYEDIYDAK 71
PE L+V NL + +TSE + +F +YG + R+ G +RG FV Y + +
Sbjct: 2 PETEHKLFVGNLSWTVTSESLIQVFPEYGTVVGARVLYDGETGRSRGYGFVCYSKRSELE 181
Query: 72 TAVDHLSGFNVANRYLIV 89
TA+ L+ + R + V
Sbjct: 182TALISLNNVELEGRAIRV 235
>TC16529 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(22%)
Length = 761
Score = 40.8 bits (94), Expect = 7e-05
Identities = 24/78 (30%), Positives = 42/78 (53%), Gaps = 3/78 (3%)
Frame = +1
Query: 17 VNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTAFVVYEDIYDAKTA 73
V LYV +L N+ ++YD+F + G + +R+ + TR G +V + + DA A
Sbjct: 325 VTTSLYVGDLDQNVNDSQLYDLFNQVGQVVSVRVCRDLTTRRSLGYGYVNFSNPPDAARA 504
Query: 74 VDHLSGFNVANRYLIVLY 91
+D L+ + NR + V+Y
Sbjct: 505 LDVLNFTPLNNRPIRVMY 558
>BE122637
Length = 332
Score = 40.4 bits (93), Expect = 9e-05
Identities = 24/79 (30%), Positives = 42/79 (52%), Gaps = 3/79 (3%)
Frame = +2
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNK---DTRGTAFVVYEDIYDAKTAVDHL 77
L VRN+P + ++E+ F ++G +R + I + + RG AFV + D YDA A H+
Sbjct: 29 LLVRNIPLDCRADEIRVPFERFGPVRDVYIPKDYYSGEPRGFAFVQFVDPYDASEAQYHM 208
Query: 78 SGFNVANRYLIVLYYQQAK 96
+ A R + V+ + +
Sbjct: 209 NRQIFAGREITVVVASETR 265
>TC8818 weakly similar to PIR|T48456|T48456 rna binding protein-like -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(90%)
Length = 1079
Score = 40.0 bits (92), Expect = 1e-04
Identities = 21/64 (32%), Positives = 33/64 (50%), Gaps = 3/64 (4%)
Frame = +3
Query: 20 VLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDAKTAVDH 76
VLYV +P +E+ FG++G I+++RI NK T R F+ +E AK D
Sbjct: 216 VLYVGRIPHGFYEKEIEGYFGQFGTIKRLRIARNKKTGKSRHFGFIEFESAEVAKIVADT 395
Query: 77 LSGF 80
+ +
Sbjct: 396 MHNY 407
>AV417836
Length = 429
Score = 38.9 bits (89), Expect = 3e-04
Identities = 23/77 (29%), Positives = 39/77 (49%), Gaps = 3/77 (3%)
Frame = +1
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDAKTAVDHL 77
L VRNLP + E++ F +YG ++ + + N T RG FV Y DA A HL
Sbjct: 127 LLVRNLPLDARPEDLRGPFERYGPVKDVYLPKNYYTGEPRGFGFVKYRFGEDAAEAKHHL 306
Query: 78 SGFNVANRYLIVLYYQQ 94
+ + R + +++ ++
Sbjct: 307 NHTIIGGREIRIVFAEE 357
>TC14532 similar to UP|Q9FYA8 (Q9FYA8) SC35-like splicing factor SCL30a, 30a
kD, partial (71%)
Length = 1073
Score = 38.9 bits (89), Expect = 3e-04
Identities = 28/106 (26%), Positives = 52/106 (48%), Gaps = 3/106 (2%)
Frame = +3
Query: 14 PPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDA 70
P ++ L VRNL + E++ FG++G ++ I + + T RG FV + D DA
Sbjct: 153 PRDLPTSLLVRNLRHDCRPEDLRRPFGQFGPLKDIYLPKDYYTGQPRGFGFVQFVDPADA 332
Query: 71 KTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEK 116
A H+ G + R L V++ ++ + K + + +E R ++
Sbjct: 333 ADAKYHMDGQILLGRELTVVFAEENR-KKPTEMRHRERSRGRSYDR 467
>AV411398
Length = 427
Score = 37.4 bits (85), Expect = 8e-04
Identities = 21/78 (26%), Positives = 38/78 (47%), Gaps = 2/78 (2%)
Frame = +3
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT--RGTAFVVYEDIYDAKTAVDHLS 78
L +N+P+ T E++ +F +YG + ++ + T RG AFV +A A++ L
Sbjct: 84 LVAQNVPWTSTPEDVRSLFERYGTVLEVELSMYNKTRSRGLAFVEMSSPEEALEALNKLE 263
Query: 79 GFNVANRYLIVLYYQQAK 96
+ R L + Y + K
Sbjct: 264 SYEFEGRVLKLNYARPKK 317
>TC13677 similar to UP|Q8H0P8 (Q8H0P8) RNA-binding protein, partial (41%)
Length = 577
Score = 37.0 bits (84), Expect = 0.001
Identities = 24/76 (31%), Positives = 36/76 (46%), Gaps = 6/76 (7%)
Frame = +2
Query: 13 LPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNK------DTRGTAFVVYED 66
LPP+ LYV LP + T E+ IF + R++R+ T + D FV + +
Sbjct: 20 LPPDA*STLYVEGLPSDCTKREVAHIFRPFVGYREVRLVTKESKHRGGDPLILCFVDFAN 199
Query: 67 IYDAKTAVDHLSGFNV 82
A TA+ L G+ V
Sbjct: 200PACAATALSALQGYKV 247
>BP041539
Length = 521
Score = 35.4 bits (80), Expect = 0.003
Identities = 20/74 (27%), Positives = 40/74 (54%), Gaps = 3/74 (4%)
Frame = +2
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTAFVVYEDIYDAKTAVDHL 77
LYV +L I ++YD+F + G + +R+ + T+ G +V + + DA TA+D L
Sbjct: 293 LYVGDLDTEINDSQLYDLFNQIGQVVSVRVCRDLATQQSLGYGYVNFTNPKDAATALDVL 472
Query: 78 SGFNVANRYLIVLY 91
+ + + + ++Y
Sbjct: 473 NFTPLNGKPIRIMY 514
>TC9521 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (56%)
Length = 598
Score = 35.0 bits (79), Expect = 0.004
Identities = 27/81 (33%), Positives = 37/81 (45%)
Frame = +1
Query: 9 GNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIY 68
GNTRL YV L S ++ +F +YG +R + + R AFV + D
Sbjct: 85 GNTRL--------YVGRLSSRTRSRDLERVFSRYGRVRDVDM-----KRDYAFVEFSDPR 225
Query: 69 DAKTAVDHLSGFNVANRYLIV 89
DA A +L G +V LIV
Sbjct: 226 DADDARYNLDGRDVDGSRLIV 288
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,376,454
Number of Sequences: 28460
Number of extensions: 11860
Number of successful extensions: 94
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 90
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 91
length of query: 124
length of database: 4,897,600
effective HSP length: 79
effective length of query: 45
effective length of database: 2,649,260
effective search space: 119216700
effective search space used: 119216700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 50 (23.9 bits)
Medicago: description of AC146941.10


