

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146866.3 + phase: 0
(482 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
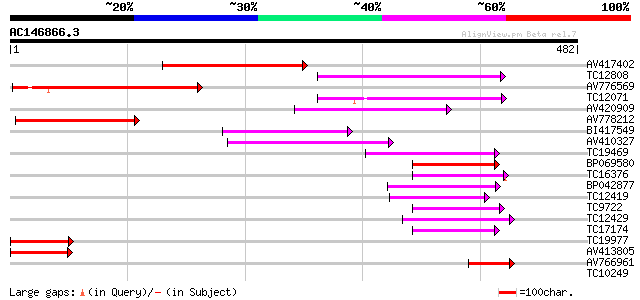
Score E
Sequences producing significant alignments: (bits) Value
AV417402 171 2e-43
TC12808 weakly similar to GB|CAA16564.1|2827556|ATT12H17 predict... 111 2e-25
AV776569 94 5e-20
TC12071 weakly similar to GB|AAP31968.1|30387605|BT006624 At2g04... 93 1e-19
AV420909 80 7e-16
AV778212 77 5e-15
BI417549 73 9e-14
AV410327 73 9e-14
TC19469 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, par... 68 3e-12
BP069580 66 1e-11
TC16376 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (17%) 60 1e-09
BP042877 60 1e-09
TC12419 58 4e-09
TC9722 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (16%) 57 7e-09
TC12429 similar to UP|Q9FKQ1 (Q9FKQ1) Emb|CAB89401.1 (AT5g65380/... 57 7e-09
TC17174 weakly similar to UP|Q940N9 (Q940N9) AT4g21910/T8O5_120,... 55 3e-08
TC19977 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (20%) 49 2e-06
AV413805 48 3e-06
AV766961 42 3e-04
TC10249 similar to UP|Q84LL9 (Q84LL9) Salt tolerance protein 3, ... 31 0.51
>AV417402
Length = 371
Score = 171 bits (433), Expect = 2e-43
Identities = 72/123 (58%), Positives = 99/123 (79%)
Frame = +3
Query: 131 LTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLYFSSVYKKSWI 190
LTYC+ +S+LLHIP+N+LLV +GI G+++ V TN NLVI L +++ S ++KK+W
Sbjct: 3 LTYCAVLSILLHIPINYLLVSVLHLGIRGIALGAVWTNFNLVISLIIYIWVSGIHKKTWT 182
Query: 191 SPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIASMGILIQTTSL 250
S C KGW +LL+LAIP+C+SVCLEWWWYE MI++CGLL+NP AT+ASMG+LIQTT+L
Sbjct: 183 GISSACFKGWKALLNLAIPSCISVCLEWWWYEIMILLCGLLINPHATVASMGVLIQTTAL 362
Query: 251 VYV 253
+Y+
Sbjct: 363 IYI 371
>TC12808 weakly similar to GB|CAA16564.1|2827556|ATT12H17 predicted protein
{Arabidopsis thaliana;}, partial (32%)
Length = 624
Score = 111 bits (278), Expect = 2e-25
Identities = 57/160 (35%), Positives = 89/160 (55%)
Frame = +1
Query: 262 VSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELT 321
VSTR+ NELGAN+ +A S VSL L ++LG L R WG F++DR I+
Sbjct: 1 VSTRVSNELGANQAGRAYRSACVSLALGLILGCIGSLVMVAARGIWGPLFSHDRGIINGV 180
Query: 322 SIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGF 381
+ ++ L E+ N P G++RG+ARP +G NLG FY + +P+ + F LG
Sbjct: 181 KKTMLLMALVEVFNFPLAVCGGIVRGTARPWLGMYANLGGFYFLALPLGVVFAFKLNLGL 360
Query: 382 PGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELTKS 421
GL+IGL+ +C L+++ + R +W + +A+ L +
Sbjct: 361 FGLFIGLITGIVTCLSLLVIFIARINWEEEASKAQALASN 480
>AV776569
Length = 633
Score = 94.0 bits (232), Expect = 5e-20
Identities = 52/164 (31%), Positives = 102/164 (61%), Gaps = 2/164 (1%)
Frame = +2
Query: 3 SMIFLGYLGEMELAGGSLSIGFANITGYS--VISGLAMGMEPICGQAYGAKQWKILGLTL 60
++ F+G++GE++LA ++S+ + I G+S V+ G+ +E +CGQA+GA Q ++LG+ +
Sbjct: 137 TLTFVGHVGELDLA--AVSVENSCIAGFSFGVMLGMGSALETLCGQAFGAGQSRMLGVYM 310
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
QR+ ++L +T++ + ++ IL GQ EIS A F L+++P LF + P++
Sbjct: 311 QRSWVILFTTALILVPAYVWSPPILRVIGQTTEISEAAGKFALWMLPQLFAYAFNFPMQK 490
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAM 164
+L++Q + + SA ++LHI ++LL++ G+ G +I +
Sbjct: 491 FLQSQRKVQVMLWISATVLVLHIFFSWLLILKLGWGLTGAAITL 622
>TC12071 weakly similar to GB|AAP31968.1|30387605|BT006624 At2g04100
{Arabidopsis thaliana;}, partial (32%)
Length = 764
Score = 92.8 bits (229), Expect = 1e-19
Identities = 57/163 (34%), Positives = 90/163 (54%), Gaps = 2/163 (1%)
Frame = +3
Query: 262 VSTRIGNELGANRPQKARISMIVSLFLAMV--LGLGAMLFTTLMRNQWGKFFTNDREILE 319
VSTR+ NELGA +P++AR ++ + LA + + L + LF R+ G F+ND E++
Sbjct: 42 VSTRVSNELGAGKPKEARDAIFAVIILATLDAIILSSALFCC--RHVLGFAFSNDLEVVH 215
Query: 320 LTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKL 379
+ ++P++ L + GV RG+ IGA NL ++Y +G+PVA+ LGFV KL
Sbjct: 216 SVAKIVPLLCLSVCVDSFLGVLSGVARGAGWQKIGAVTNLLAYYAIGIPVALLLGFVLKL 395
Query: 380 GFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKELTKSS 422
GLWIG+L + +++ ++ T W Q A T S
Sbjct: 396 NAKGLWIGILTGSTTQTLVLALLTAFTKWEKQAPLATVRTSES 524
>AV420909
Length = 421
Score = 80.1 bits (196), Expect = 7e-16
Identities = 42/133 (31%), Positives = 74/133 (55%)
Frame = +3
Query: 243 ILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTTL 302
+ + TT+L Y ++ STR+ NELGA P+ A+ ++ V + + +V + F
Sbjct: 6 VCLNTTTLHYFVTYAVGAAASTRVSNELGAGNPKTAKGAVRVVVIVGIVEAIIVSAFFIC 185
Query: 303 MRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSF 362
RN G ++ND+E+++ + + P++ +G+ GV RG IGA NLG++
Sbjct: 186 FRNVLGYAYSNDKEVVDYVADMAPLLCGSVIGDSLIGALSGVARGGGFQQIGAYANLGAY 365
Query: 363 YLVGMPVAIFLGF 375
Y+VG+P+ + LGF
Sbjct: 366 YVVGIPIGLLLGF 404
>AV778212
Length = 557
Score = 77.4 bits (189), Expect = 5e-15
Identities = 34/105 (32%), Positives = 64/105 (60%)
Frame = +1
Query: 6 FLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTLQRTVL 65
F G+LG + LA +L + Y ++ G+ +E +CGQAYGA ++ +LG+ +QR +
Sbjct: 235 FAGHLGNLALAAANLGNDGIQLFAYGLMLGMGSAVETLCGQAYGAHKYDMLGIYMQRAFI 414
Query: 66 LLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLF 110
+L T IP++ +++ K+ILL G+ +++S F+ L+P ++
Sbjct: 415 VLTITGIPLTVVYVFCKQILLVLGESDDVASAGAIFVYGLIPQIY 549
>BI417549
Length = 404
Score = 73.2 bits (178), Expect = 9e-14
Identities = 32/110 (29%), Positives = 60/110 (54%)
Frame = +1
Query: 182 SSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIASM 241
SS +K+ S + G L++P+ VC++WW E ++++ GL NP+ + +
Sbjct: 10 SSACEKTRTPFSKRALSGLGEFFRLSVPSAAMVCVKWWASELLVLLAGLFSNPELETSIL 189
Query: 242 GILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMV 291
I + ++L + P + STR+ NELGA P+ AR+ + +++ LA +
Sbjct: 190 SICLTISTLHFTIPYGFGVAASTRVANELGAGNPKAARVVVSIAMSLAFI 339
>AV410327
Length = 424
Score = 73.2 bits (178), Expect = 9e-14
Identities = 35/141 (24%), Positives = 68/141 (47%)
Frame = +2
Query: 186 KKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIASMGILI 245
K++W S L+ + V +CLE W+++ ++++ GLL +P+ + S+ I
Sbjct: 2 KRTWQGFSWQAFSELPEFFKLSAASAVMLCLETWYFQILVLLAGLLPHPELALDSLSICT 181
Query: 246 QTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRN 305
+ V++ + S R+ NELGA P+ A S++V ++ ++ + A L +R+
Sbjct: 182 TISGWVFMISVGFNAAASVRVSNELGARNPKSASFSVVVVTLISFIMSVIAALVVLALRD 361
Query: 306 QWGKFFTNDREILELTSIVLP 326
FT E+ S + P
Sbjct: 362 IISYVFTGGEEVAAAVSDLCP 424
>TC19469 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, partial (26%)
Length = 501
Score = 68.2 bits (165), Expect = 3e-12
Identities = 35/114 (30%), Positives = 61/114 (52%)
Frame = +1
Query: 303 MRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSF 362
+R + FT D E+ E + P++ + L N GV G+ P++ A +N+G +
Sbjct: 7 LRERLAYVFTLDSEVAEAVGDLSPLLSISILLNSVPPVLSGVSVGAGWPSVVAYVNIGCY 186
Query: 363 YLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAK 416
YL+G+PV + LG + L G+WIG+L ++++ + +TDW QV A+
Sbjct: 187 YLIGIPVGVVLGNLIHLQEKGVWIGMLFGTFVQTVMLITITLKTDWENQVVIAR 348
>BP069580
Length = 483
Score = 65.9 bits (159), Expect = 1e-11
Identities = 30/74 (40%), Positives = 45/74 (60%)
Frame = -1
Query: 343 GVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVV 402
G RG IGA INLGS+YLVG+P+A+ L FV +G GLW+G++ A ++++
Sbjct: 261 GTARGCGLQKIGAFINLGSYYLVGIPLAMVLAFVLHIGGKGLWLGIICALIVQVFSLMII 82
Query: 403 LCRTDWNLQVQRAK 416
R DW + +R +
Sbjct: 81 TLRIDWEKEARRPR 40
>TC16376 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (17%)
Length = 550
Score = 59.7 bits (143), Expect = 1e-09
Identities = 33/85 (38%), Positives = 48/85 (55%), Gaps = 3/85 (3%)
Frame = +2
Query: 343 GVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVV 402
GV G T A +N+G +Y +G+P LGF K G G+W+G+LA ++++ V
Sbjct: 17 GVAVGCGWQTFVAYVNVGCYYGIGIPFGAVLGFYFKFGAMGIWLGMLAGTVLQTIILVWV 196
Query: 403 LCRTDWNLQVQRA-KELTK--SSTT 424
RTDWN +++ A K L K STT
Sbjct: 197 TFRTDWNKEIEEATKRLNKWEESTT 271
>BP042877
Length = 489
Score = 59.7 bits (143), Expect = 1e-09
Identities = 31/96 (32%), Positives = 52/96 (53%)
Frame = -1
Query: 322 SIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGF 381
S + P++ L L + Q GV G P A +N+G +Y VG+P+ LGF + G
Sbjct: 489 SDLCPLLALSILLHGIQPVLSGVAVGCGWPAFVAYVNVGCYYGVGIPLGSVLGFYFQFGA 310
Query: 382 PGLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAKE 417
G+W+G+L ++++ V RTDW+ +V+ A +
Sbjct: 309 QGIWLGMLGGTTMPTIILMWVTFRTDWDQEVEEAAQ 202
>TC12419
Length = 667
Score = 57.8 bits (138), Expect = 4e-09
Identities = 29/85 (34%), Positives = 45/85 (52%)
Frame = +3
Query: 324 VLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPG 383
V P++ + L + Q GV RG + A +NL +FYL+G+P++ L F L + G
Sbjct: 15 VAPLLAISILLDSIQGVLSGVARGCGWQHLAAYVNLATFYLIGLPISCLLAFKTSLQYKG 194
Query: 384 LWIGLLAAQGSCAMLMLVVLCRTDW 408
LWIGL+ A +L++ R W
Sbjct: 195 LWIGLICGMVCQAGTLLLLTTRAKW 269
>TC9722 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (16%)
Length = 571
Score = 57.0 bits (136), Expect = 7e-09
Identities = 30/79 (37%), Positives = 44/79 (54%), Gaps = 1/79 (1%)
Frame = +3
Query: 343 GVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVV 402
GV G T A +N+G +Y VG+P+ LGF K G G+W+G+L ++++ V
Sbjct: 9 GVAVGCGCQTFVAYVNVGCYYGVGIPLGAVLGFYFKFGAKGIWLGMLGGTVMQTIILMWV 188
Query: 403 LCRTDWNLQVQRA-KELTK 420
RTDW +V+ A K L K
Sbjct: 189 TFRTDWIKEVEEASKRLNK 245
>TC12429 similar to UP|Q9FKQ1 (Q9FKQ1) Emb|CAB89401.1 (AT5g65380/MNA5_11),
partial (22%)
Length = 523
Score = 57.0 bits (136), Expect = 7e-09
Identities = 34/97 (35%), Positives = 54/97 (55%), Gaps = 2/97 (2%)
Frame = +1
Query: 335 NCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGS 394
N Q GV GS + A INLG +YLVG+P+ +G+V G G+W G++ +
Sbjct: 55 NSIQPVLSGVAVGSGWQSYVAYINLGCYYLVGVPLGFIMGWVFNQGVMGIWAGMIFGGTA 234
Query: 395 CAMLML-VVLCRTDWNLQVQRAK-ELTKSSTTSDDVD 429
L+L V+ R DW+ + ++AK LTK S + +++
Sbjct: 235 LQTLILGVITIRCDWDGEAEKAKLHLTKWSDSKRELN 345
>TC17174 weakly similar to UP|Q940N9 (Q940N9) AT4g21910/T8O5_120, partial
(18%)
Length = 723
Score = 55.1 bits (131), Expect = 3e-08
Identities = 27/74 (36%), Positives = 43/74 (57%)
Frame = +1
Query: 343 GVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVV 402
GV G+ + A +NLG +Y++G+PV I LG V G+WIG+L ++++++
Sbjct: 46 GVALGAGWRSTVAYVNLGCYYIIGLPVGIVLGNVFHWQVKGIWIGMLFGTLVQTIVLVII 225
Query: 403 LCRTDWNLQVQRAK 416
RTDW QV A+
Sbjct: 226 TYRTDWEEQVTIAR 267
>TC19977 similar to UP|Q9LVD9 (Q9LVD9) Gb|AAC28507.1, partial (20%)
Length = 452
Score = 48.9 bits (115), Expect = 2e-06
Identities = 22/54 (40%), Positives = 34/54 (62%)
Frame = +3
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWK 54
M + IF G+LG +ELA SL + Y ++ G+ +E +CGQAYGAK+++
Sbjct: 291 MSTQIFSGHLGNLELAAASLGNTGIQVFAYGLMLGMGSAVETLCGQAYGAKKFE 452
>AV413805
Length = 418
Score = 48.1 bits (113), Expect = 3e-06
Identities = 18/54 (33%), Positives = 40/54 (73%), Gaps = 1/54 (1%)
Frame = +3
Query: 1 MISMIFLGYLGEM-ELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQW 53
++S++ +G+ E+ +G +++I FA +TG+SV+ G++ +E +CGQ +GA+++
Sbjct: 246 VVSLMMVGHFSELVSFSGVAIAISFAEVTGFSVLLGMSGALETLCGQTFGAEEF 407
>AV766961
Length = 283
Score = 41.6 bits (96), Expect = 3e-04
Identities = 20/39 (51%), Positives = 25/39 (63%)
Frame = -2
Query: 391 AQGSCAMLMLVVLCRTDWNLQVQRAKELTKSSTTSDDVD 429
AQ +C + MLVVL RT+W Q QRAKELT + + D
Sbjct: 282 AQAACMVTMLVVLARTNWEGQAQRAKELTSLDSGEEGGD 166
>TC10249 similar to UP|Q84LL9 (Q84LL9) Salt tolerance protein 3, partial
(35%)
Length = 1096
Score = 30.8 bits (68), Expect = 0.51
Identities = 26/82 (31%), Positives = 44/82 (52%)
Frame = -2
Query: 113 SILHPLRIYLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLV 172
S +HP T ITLP + VS+ + IPL +++ + I ++ ++L+L+
Sbjct: 381 SRVHPPSTISITSNITLPRPFTV*VSISIPIPLPISILITMLLPIT*IT-----SSLSLI 217
Query: 173 ILLSSFLYFSSVYKKSWISPSL 194
+LL L FSS++ S+ S SL
Sbjct: 216 LLL---LPFSSLFCLSFPS*SL 160
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.141 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,233,880
Number of Sequences: 28460
Number of extensions: 147680
Number of successful extensions: 1338
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 1317
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1336
length of query: 482
length of database: 4,897,600
effective HSP length: 94
effective length of query: 388
effective length of database: 2,222,360
effective search space: 862275680
effective search space used: 862275680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146866.3


