

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146863.3 + phase: 0
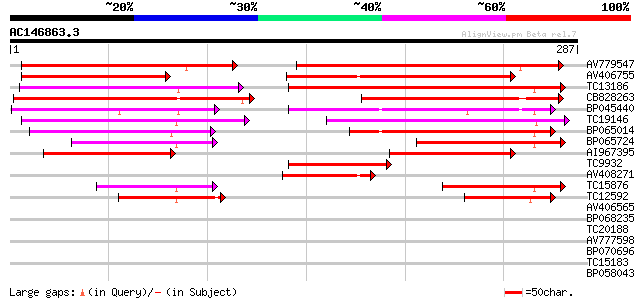
(287 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV779547 144 2e-35
AV406755 125 6e-30
TC13186 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candidat... 113 4e-26
CB828263 105 7e-24
BP045440 100 4e-22
TC19146 similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-contain... 87 3e-18
BP065014 85 2e-17
BP065724 64 3e-11
AI967395 62 9e-11
TC9932 weakly similar to UP|Q93YA7 (Q93YA7) Resistance gene-like... 57 4e-09
AV408271 50 3e-07
TC15876 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-... 47 4e-06
TC12592 44 4e-05
AV406565 39 0.001
BP068235 35 0.011
TC20188 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-... 35 0.015
AV777598 34 0.033
BP070696 31 0.28
TC15183 similar to PIR|T49142|T49142 CCR4-associated factor 1-li... 28 1.4
BP058043 27 5.2
>AV779547
Length = 556
Score = 144 bits (363), Expect = 2e-35
Identities = 77/137 (56%), Positives = 93/137 (67%), Gaps = 2/137 (1%)
Frame = +1
Query: 146 VSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLYIVVFSK 205
VSFRG DTR NFTDHL AL +G F+DDT L+KG+ I+ L +AIE SQ+ IVVFSK
Sbjct: 109 VSFRGEDTRNNFTDHLLGALYGKGFVTFKDDTMLRKGKNISTELIQAIEGSQILIVVFSK 288
Query: 206 NYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSKH--GFKHGL 263
YASSTWCL+EL I + +LP+ DV PSEV+KQSG YGEA KH FK L
Sbjct: 289 YYASSTWCLQELAKIADGIVGKRQTVLPVVCDVTPSEVRKQSGNYGEAFLKHEERFKEDL 468
Query: 264 NMVHRWRETLTQVGNIS 280
MV RWR+ L +V ++S
Sbjct: 469 EMVQRWRKALAEVADLS 519
Score = 100 bits (248), Expect = 4e-22
Identities = 55/113 (48%), Positives = 72/113 (63%), Gaps = 4/113 (3%)
Frame = +1
Query: 7 GDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPS 66
G + + AIE SQ+ IVVFSK +A S +CL ELA I V + +LP+ DV PS
Sbjct: 217 GKNISTELIQAIEGSQILIVVFSKYYASSTWCLQELAKIADGIVGKRQTVLPVVCDVTPS 396
Query: 67 EVRKQSGGYGESLAKLEEIAPQ----VQRWREALQLVGNISGWDLCHKPQHAE 115
EVRKQSG YGE+ K EE + VQRWR+AL V ++SGWD+ +KP+H +
Sbjct: 397 EVRKQSGNYGEAFLKHEERFKEDLEMVQRWRKALAEVADLSGWDVTNKPEHEQ 555
>AV406755
Length = 425
Score = 125 bits (315), Expect = 6e-30
Identities = 57/116 (49%), Positives = 84/116 (72%)
Frame = +3
Query: 141 NYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLYI 200
+YDVF+SFRG ++R +FT HL+ AL+ GI F D+ +L++GE I+ L +AIE S++ I
Sbjct: 81 SYDVFLSFRGEESRRSFTSHLYTALKNAGIKVFMDN-ELQRGEDISSSLLKAIEDSRIAI 257
Query: 201 VVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSK 256
++FS NY S WCL ELE I+ C + G+ ++P+FY+VDPS+++KQ G GEA K
Sbjct: 258 IIFSTNYTGSKWCLDELEKIIECQRTIGQEVMPVFYNVDPSDIRKQRGTVGEAFRK 425
Score = 70.9 bits (172), Expect = 2e-13
Identities = 32/75 (42%), Positives = 49/75 (64%)
Frame = +3
Query: 7 GDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPS 66
G+ + AIEDS++ I++FS N+ S +CL EL I+ C G+ ++P+FY+VDPS
Sbjct: 201 GEDISSSLLKAIEDSRIAIIIFSTNYTGSKWCLDELEKIIECQRTIGQEVMPVFYNVDPS 380
Query: 67 EVRKQSGGYGESLAK 81
++RKQ G GE+ K
Sbjct: 381 DIRKQRGTVGEAFRK 425
>TC13186 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candidate
resistance protein KR1, partial (6%)
Length = 586
Score = 113 bits (282), Expect = 4e-26
Identities = 60/142 (42%), Positives = 90/142 (63%), Gaps = 2/142 (1%)
Frame = +1
Query: 142 YDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLYIV 201
YDVF+S DTRF F+ L+ AL +GI F DD + + ++ +AIE+S++ IV
Sbjct: 103 YDVFISLITSDTRFGFSGDLYTALSDKGILTFIDDGLPRPDDIMSTVNSKAIESSRIAIV 282
Query: 202 VFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSKHGFKH 261
V S+NYASS++CL EL YIL ++ G I P+FY V+PS+V+ Q GG+G+A + H +
Sbjct: 283 VISENYASSSFCLDELMYILKRREEKGLLIWPVFYQVEPSDVRYQKGGFGKAFASHEERF 462
Query: 262 GLN--MVHRWRETLTQVGNISS 281
G + V +WR L QV ++ +
Sbjct: 463 GSDSAKVRKWRNALKQVADLKA 528
Score = 84.7 bits (208), Expect = 2e-17
Identities = 47/117 (40%), Positives = 67/117 (57%), Gaps = 4/117 (3%)
Frame = +1
Query: 6 PGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDP 65
P D++ AIE S++ IVV S+N+A S FCL EL YIL G I P+FY V+P
Sbjct: 220 PDDIMSTVNSKAIESSRIAIVVISENYASSSFCLDELMYILKRREEKGLLIWPVFYQVEP 399
Query: 66 SEVRKQSGGYGESLAKLEE----IAPQVQRWREALQLVGNISGWDLCHKPQHAELEN 118
S+VR Q GG+G++ A EE + +V++WR AL+ V ++ + H L N
Sbjct: 400 SDVRYQKGGFGKAFASHEERFGSDSAKVRKWRNALKQVADLKAFHFFKHGYHTSLIN 570
>CB828263
Length = 570
Score = 105 bits (263), Expect = 7e-24
Identities = 49/102 (48%), Positives = 70/102 (68%)
Frame = +2
Query: 179 LKKGEFIAPGLFRAIEASQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDV 238
L++G+ I+ L RAIE ++L ++VFSKNY +S WCL EL IL C + G+ +LP+FYD+
Sbjct: 8 LQRGDEISSSLLRAIEEAKLSVIVFSKNYGNSKWCLDELVKILECKRTRGQIVLPVFYDI 187
Query: 239 DPSEVQKQSGGYGEALSKHGFKHGLNMVHRWRETLTQVGNIS 280
DPS V+ Q+G Y EA KHG ++ V +WRE L + N+S
Sbjct: 188 DPSHVRNQTGTYAEAFVKHG---QVDKVQKWREALREAANLS 304
Score = 102 bits (254), Expect = 8e-23
Identities = 50/127 (39%), Positives = 79/127 (61%), Gaps = 5/127 (3%)
Frame = +2
Query: 3 DYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYD 62
D GD + AIE++++ ++VFSKN+ +S +CL EL IL C G+ +LP+FYD
Sbjct: 5 DLQRGDEISSSLLRAIEEAKLSVIVFSKNYGNSKWCLDELVKILECKRTRGQIVLPVFYD 184
Query: 63 VDPSEVRKQSGGYGESLAKLEEIAPQVQRWREALQLVGNISGWDLCHKPQHAEL-----E 117
+DPS VR Q+G Y E+ K ++ +VQ+WREAL+ N+SGWD +E+ +
Sbjct: 185 IDPSHVRNQTGTYAEAFVKHGQV-DKVQKWREALREAANLSGWDCSVNRMESEVIEKIAK 361
Query: 118 NIIEHIN 124
+++E +N
Sbjct: 362 DVLEKLN 382
>BP045440
Length = 535
Score = 100 bits (248), Expect = 4e-22
Identities = 57/139 (41%), Positives = 82/139 (58%), Gaps = 4/139 (2%)
Frame = -3
Query: 142 YDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLYIV 201
Y +F+SFRG DTR +FT L+ L R G F+DD L+ G I L IE S+ I+
Sbjct: 527 YQIFISFRGMDTRDDFTKPLYQGLCREGFVTFKDDESLEGGSPIEK-LLDDIEESRFAII 351
Query: 202 VFSKNYASSTWCLRELEYILHCSKKYGKH--ILPIFYDVDPSEVQKQSGGYGEALSKHGF 259
+ SKNYA S WCL+EL IL C + GK+ +LP +V P+EV+ Y +A++KH
Sbjct: 350 ILSKNYAESEWCLKELSKILECKDQEGKNQLVLPSSTEVTPTEVRHVINRYADAMAKHE- 174
Query: 260 KHGLN--MVHRWRETLTQV 276
K+G++ + W++ L V
Sbjct: 173 KNGIDSKTIQAWKKALFDV 117
Score = 62.4 bits (150), Expect = 9e-11
Identities = 39/110 (35%), Positives = 63/110 (56%), Gaps = 5/110 (4%)
Frame = -3
Query: 2 DDYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGK--CILPI 59
D+ G I K IE+S+ I++ SKN+A+S +CL EL+ IL C GK +LP
Sbjct: 425 DESLEGGSPIEKLLDDIEESRFAIIILSKNYAESEWCLKELSKILECKDQEGKNQLVLPS 246
Query: 60 FYDVDPSEVRKQSGGYGESLAKLEE---IAPQVQRWREALQLVGNISGWD 106
+V P+EVR Y +++AK E+ + +Q W++AL V ++G++
Sbjct: 245 STEVTPTEVRHVINRYADAMAKHEKNGIDSKTIQAWKKALFDVCTLTGFE 96
>TC19146 similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-containing protein
TSDC, partial (12%)
Length = 701
Score = 87.4 bits (215), Expect = 3e-18
Identities = 48/125 (38%), Positives = 67/125 (53%), Gaps = 2/125 (1%)
Frame = +1
Query: 161 LFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLYIVVFSKNYASSTWCLRELEYI 220
L+ AL+R G F DD L G IA L A E S+L I+VFS+NY S+WCL EL I
Sbjct: 1 LYHALRREGFKTFMDDEGLVGGNEIAKSLLDASEKSRLSIIVFSENYGYSSWCLDELVKI 180
Query: 221 LHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSKHGFKHGLNM--VHRWRETLTQVGN 278
+ C + + + PIFY V+ S+V Q Y A+ H +G + V +WR L+ +
Sbjct: 181 VECMETKKQLVWPIFYKVEKSDVTSQENSYEHAMIGHEKNYGKDSDEVKKWRSALSAMAK 360
Query: 279 ISSSN 283
+ N
Sbjct: 361 LEGHN 375
Score = 61.2 bits (147), Expect = 2e-10
Identities = 35/120 (29%), Positives = 66/120 (54%), Gaps = 5/120 (4%)
Frame = +1
Query: 7 GDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPS 66
G+ + + A E S++ I+VFS+N+ S +CL EL I+ C + + PIFY V+ S
Sbjct: 64 GNEIAKSLLDASEKSRLSIIVFSENYGYSSWCLDELVKIVECMETKKQLVWPIFYKVEKS 243
Query: 67 EVRKQSGGYGESLAKLE----EIAPQVQRWREALQLVGNISGWDLC-HKPQHAELENIIE 121
+V Q Y ++ E + + +V++WR AL + + G ++ +K ++ ++ I+E
Sbjct: 244 DVTSQENSYEHAMIGHEKNYGKDSDEVKKWRSALSAMAKLEGHNVKENKYEYEFIKKIVE 423
>BP065014
Length = 498
Score = 84.7 bits (208), Expect = 2e-17
Identities = 46/106 (43%), Positives = 65/106 (60%), Gaps = 2/106 (1%)
Frame = -2
Query: 173 FRDDTKLKKGEFIAPGLFRAIEASQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHIL 232
F+DD L+ G I L AIE S++ IVV S+N+A S WCL+EL IL C KK +L
Sbjct: 497 FKDDLSLEDGAPIEE-LLGAIEESRVAIVVLSENFADSKWCLKELVKILDCRKKKKPLVL 321
Query: 233 PIFYDVDPSEVQKQSGGYGEALSKHGFKHGLN--MVHRWRETLTQV 276
PIFY V+P+ ++ G YGEA+++H K G + V W + L+ +
Sbjct: 320 PIFYKVEPTNIRHLKGRYGEAMAEHEKKLGKDSQTVREWTQALSDI 183
Score = 75.9 bits (185), Expect = 8e-15
Identities = 42/98 (42%), Positives = 59/98 (59%), Gaps = 4/98 (4%)
Frame = -2
Query: 11 IRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPSEVRK 70
I + AIE+S+V IVV S+NFADS +CL EL IL C +LPIFY V+P+ +R
Sbjct: 461 IEELLGAIEESRVAIVVLSENFADSKWCLKELVKILDCRKKKKPLVLPIFYKVEPTNIRH 282
Query: 71 QSGGYGESLA----KLEEIAPQVQRWREALQLVGNISG 104
G YGE++A KL + + V+ W +AL + + G
Sbjct: 281 LKGRYGEAMAEHEKKLGKDSQTVREWTQALSDISYLKG 168
>BP065724
Length = 495
Score = 63.9 bits (154), Expect = 3e-11
Identities = 30/77 (38%), Positives = 49/77 (62%), Gaps = 2/77 (2%)
Frame = -1
Query: 207 YASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSKHGFKHGLN-- 264
YA S+WCL E+ IL C +K + + PIFY V+PS+V+ Q Y +A+ K ++G +
Sbjct: 495 YADSSWCLEEVVKILECKQKNDQLVWPIFYKVEPSDVRHQRNSYEKAMIKQETRYGKDSE 316
Query: 265 MVHRWRETLTQVGNISS 281
V +WR +L+QV ++ +
Sbjct: 315 KVKKWRSSLSQVCDLKA 265
Score = 53.9 bits (128), Expect = 3e-08
Identities = 27/78 (34%), Positives = 46/78 (58%), Gaps = 4/78 (5%)
Frame = -1
Query: 32 FADSCFCLVELAYILHCSVLYGKCILPIFYDVDPSEVRKQSGGYGESLAKLE----EIAP 87
+ADS +CL E+ IL C + + PIFY V+PS+VR Q Y +++ K E + +
Sbjct: 495 YADSSWCLEEVVKILECKQKNDQLVWPIFYKVEPSDVRHQRNSYEKAMIKQETRYGKDSE 316
Query: 88 QVQRWREALQLVGNISGW 105
+V++WR +L V ++ +
Sbjct: 315 KVKKWRSSLSQVCDLKAF 262
>AI967395
Length = 205
Score = 62.4 bits (150), Expect = 9e-11
Identities = 28/67 (41%), Positives = 46/67 (67%)
Frame = +3
Query: 18 IEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDPSEVRKQSGGYGE 77
I++S++ IVVFS+++A S CL EL I+ ++ + + P++Y V+ E+R Q G YGE
Sbjct: 3 IQESRLIIVVFSEHYAVSASCLNELVNIVDVMIMNNQLVWPVYYGVEAPEIRHQRGRYGE 182
Query: 78 SLAKLEE 84
++AK EE
Sbjct: 183AMAKFEE 203
Score = 54.3 bits (129), Expect = 2e-08
Identities = 27/64 (42%), Positives = 41/64 (63%)
Frame = +3
Query: 193 IEASQLYIVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGE 252
I+ S+L IVVFS++YA S CL EL I+ + + P++Y V+ E++ Q G YGE
Sbjct: 3 IQESRLIIVVFSEHYAVSASCLNELVNIVDVMIMNNQLVWPVYYGVEAPEIRHQRGRYGE 182
Query: 253 ALSK 256
A++K
Sbjct: 183 AMAK 194
>TC9932 weakly similar to UP|Q93YA7 (Q93YA7) Resistance gene-like, partial
(5%)
Length = 689
Score = 57.0 bits (136), Expect = 4e-09
Identities = 25/52 (48%), Positives = 34/52 (65%)
Frame = -2
Query: 142 YDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAI 193
YDVF++FRG DTR +F HL+ AL G+N F DD L++G + P L + I
Sbjct: 202 YDVFINFRGKDTRHSFVSHLYTALSNAGVNVFLDDENLERGLELEPELQKLI 47
>AV408271
Length = 409
Score = 50.4 bits (119), Expect = 3e-07
Identities = 27/47 (57%), Positives = 32/47 (67%)
Frame = +1
Query: 139 EINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFI 185
E YDVFVSFRG D R F HL A +++ INAF DD KLK+G+ I
Sbjct: 271 EEKYDVFVSFRGEDIRDGFLSHLADAFRQKKINAFMDD-KLKRGQEI 408
>TC15876 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-containing
protein TSDC, partial (13%)
Length = 558
Score = 47.0 bits (110), Expect = 4e-06
Identities = 23/64 (35%), Positives = 40/64 (61%), Gaps = 2/64 (3%)
Frame = +1
Query: 220 ILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSKHGFKHGLN--MVHRWRETLTQVG 277
IL C +K + + PIFY V+PS+V+ Q Y +A+ K ++G + V +WR +L+QV
Sbjct: 16 ILECXQKNDQLVWPIFYKVEPSDVRHQRNSYEKAMIKPETRYGKDSEKVKKWRSSLSQVC 195
Query: 278 NISS 281
++ +
Sbjct: 196 DLKA 207
Score = 41.2 bits (95), Expect = 2e-04
Identities = 21/65 (32%), Positives = 37/65 (56%), Gaps = 4/65 (6%)
Frame = +1
Query: 45 ILHCSVLYGKCILPIFYDVDPSEVRKQSGGYGESLAKLE----EIAPQVQRWREALQLVG 100
IL C + + PIFY V+PS+VR Q Y +++ K E + + +V++WR +L V
Sbjct: 16 ILECXQKNDQLVWPIFYKVEPSDVRHQRNSYEKAMIKPETRYGKDSEKVKKWRSSLSQVC 195
Query: 101 NISGW 105
++ +
Sbjct: 196 DLKAF 210
>TC12592
Length = 534
Score = 43.5 bits (101), Expect = 4e-05
Identities = 18/58 (31%), Positives = 38/58 (65%), Gaps = 4/58 (6%)
Frame = +3
Query: 56 ILPIFYDVDPSEVRKQSGGYGESLAKLE----EIAPQVQRWREALQLVGNISGWDLCH 109
+LP+FY V+P+++R G +GE++A++E + + + W++AL+ + + G + CH
Sbjct: 9 VLPVFYGVEPTDIRNLKGKFGEAMAEIENKFGKDSNTIHEWKQALRDISYLKG-EPCH 179
Score = 42.0 bits (97), Expect = 1e-04
Identities = 16/48 (33%), Positives = 31/48 (64%), Gaps = 2/48 (4%)
Frame = +3
Query: 231 ILPIFYDVDPSEVQKQSGGYGEALSKHGFKHG--LNMVHRWRETLTQV 276
+LP+FY V+P++++ G +GEA+++ K G N +H W++ L +
Sbjct: 9 VLPVFYGVEPTDIRNLKGKFGEAMAEIENKFGKDSNTIHEWKQALRDI 152
>AV406565
Length = 207
Score = 38.9 bits (89), Expect = 0.001
Identities = 21/56 (37%), Positives = 33/56 (58%)
Frame = +2
Query: 130 FSSRTKDLVEINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFI 185
+S+ + + +DVF+SFRG DTR+ T HL A L R + + D L++G+ I
Sbjct: 41 WSTTSSSAPQQKHDVFLSFRGEDTRYTCTGHLQATLTRLQVKTY-IDYDLQRGDEI 205
>BP068235
Length = 511
Score = 35.4 bits (80), Expect = 0.011
Identities = 18/63 (28%), Positives = 32/63 (50%), Gaps = 5/63 (7%)
Frame = -1
Query: 39 LVELAYILHCSVLYGKCILPIFYDVDPSEVRKQSGGYGESLA-----KLEEIAPQVQRWR 93
L+ L+ L C + + + P Y V+PS++R + YG +LA + ++Q W+
Sbjct: 475 LMNLSRFLECKKMKNQLLWPCPYKVEPSDIRYRPSSYGTALA*TRTDAFGNDSEKIQTWK 296
Query: 94 EAL 96
AL
Sbjct: 295 SAL 287
Score = 32.3 bits (72), Expect = 0.096
Identities = 17/63 (26%), Positives = 28/63 (43%), Gaps = 3/63 (4%)
Frame = -1
Query: 214 LRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALS---KHGFKHGLNMVHRWR 270
L L L C K + + P Y V+PS+++ + YG AL+ F + + W+
Sbjct: 475 LMNLSRFLECKKMKNQLLWPCPYKVEPSDIRYRPSSYGTALA*TRTDAFGNDSEKIQTWK 296
Query: 271 ETL 273
L
Sbjct: 295 SAL 287
>TC20188 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-containing
protein TSDC, partial (11%)
Length = 494
Score = 35.0 bits (79), Expect = 0.015
Identities = 15/48 (31%), Positives = 26/48 (53%), Gaps = 2/48 (4%)
Frame = +1
Query: 231 ILPIFYDVDPSEVQKQSGGYGEALSKH--GFKHGLNMVHRWRETLTQV 276
+ PIFY++ S+V Q+ YG A+ H F V +W+ L+++
Sbjct: 10 VWPIFYEISXSDVSNQTKSYGHAMLAHENSFGKDSEKVQKWKSALSEM 153
Score = 33.9 bits (76), Expect = 0.033
Identities = 16/53 (30%), Positives = 30/53 (56%), Gaps = 4/53 (7%)
Frame = +1
Query: 56 ILPIFYDVDPSEVRKQSGGYGESLAKLE----EIAPQVQRWREALQLVGNISG 104
+ PIFY++ S+V Q+ YG ++ E + + +VQ+W+ AL + + G
Sbjct: 10 VWPIFYEISXSDVSNQTKSYGHAMLAHENSFGKDSEKVQKWKSALSEMAFLQG 168
>AV777598
Length = 371
Score = 33.9 bits (76), Expect = 0.033
Identities = 14/21 (66%), Positives = 18/21 (85%)
Frame = +3
Query: 142 YDVFVSFRGPDTRFNFTDHLF 162
YDVF+SF G DTRF+FT +L+
Sbjct: 309 YDVFLSFCGVDTRFSFTGNLY 371
>BP070696
Length = 429
Score = 30.8 bits (68), Expect = 0.28
Identities = 17/58 (29%), Positives = 28/58 (47%), Gaps = 2/58 (3%)
Frame = -1
Query: 221 LHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSKHGFKHG--LNMVHRWRETLTQV 276
L C KK I PIF +++ ++ Q Y + + H K+G + V +W+ L V
Sbjct: 429 LECKKKKNNIIRPIFQNMELLKIIYQINSYDDVMVAHERKNGKESDRVKKWKSALCYV 256
>TC15183 similar to PIR|T49142|T49142 CCR4-associated factor 1-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(31%)
Length = 446
Score = 28.5 bits (62), Expect = 1.4
Identities = 13/20 (65%), Positives = 16/20 (80%)
Frame = -3
Query: 66 SEVRKQSGGYGESLAKLEEI 85
SEVR+ SGG GES A L+E+
Sbjct: 384 SEVREVSGGVGESEANLDEV 325
>BP058043
Length = 375
Score = 26.6 bits (57), Expect = 5.2
Identities = 12/29 (41%), Positives = 16/29 (54%)
Frame = -3
Query: 142 YDVFVSFRGPDTRFNFTDHLFAALQRRGI 170
YDVF+S RG R F + L ++GI
Sbjct: 154 YDVFISLRGEVLRDTFIECFLKELMQKGI 68
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.140 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,674,972
Number of Sequences: 28460
Number of extensions: 81640
Number of successful extensions: 456
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 413
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 449
length of query: 287
length of database: 4,897,600
effective HSP length: 89
effective length of query: 198
effective length of database: 2,364,660
effective search space: 468202680
effective search space used: 468202680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146863.3


