

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146856.6 - phase: 0 /pseudo
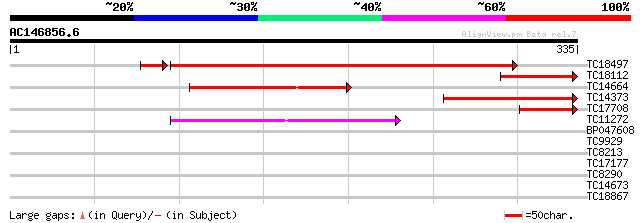
(335 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18497 similar to UP|Q9FJX5 (Q9FJX5) Genomic DNA, chromosome 5,... 381 e-108
TC18112 similar to UP|Q9FJX5 (Q9FJX5) Genomic DNA, chromosome 5,... 102 7e-23
TC14664 weakly similar to AAR24650 (AAR24650) At5g63160, partial... 93 5e-20
TC14373 weakly similar to UP|Q9FJX5 (Q9FJX5) Genomic DNA, chromo... 90 5e-19
TC17708 similar to UP|Q9FJX5 (Q9FJX5) Genomic DNA, chromosome 5,... 74 4e-14
TC11272 similar to UP|Q8LIQ8 (Q8LIQ8) OJ1370_E02.2 protein (OJ13... 41 3e-04
BP047608 32 0.20
TC9929 similar to UP|O81396 (O81396) Aconitase-iron regulated pr... 28 2.2
TC8213 28 2.8
TC17177 homologue to UP|GAPN_PEA (P81406) NADP-dependent glycera... 27 4.8
TC8290 27 6.3
TC14673 similar to UP|Q9LH76 (Q9LH76) Genomic DNA, chromosome 3,... 26 8.2
TC18867 similar to UP|Q9ASV2 (Q9ASV2) At1g06730/F4H5_22, partial... 26 8.2
>TC18497 similar to UP|Q9FJX5 (Q9FJX5) Genomic DNA, chromosome 5, TAC
clone:K9I9 (AT5g67480/K9I9_4), partial (61%)
Length = 696
Score = 381 bits (979), Expect(2) = e-108
Identities = 183/205 (89%), Positives = 194/205 (94%)
Frame = +2
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
MLKQA R NRWRSISIFGVPHD VRVFIR+LYSS EKEEM+E++LHLLVLSHVY VP L
Sbjct: 80 MLKQAKRHNRWRSISIFGVPHDVVRVFIRFLYSSRLEKEEMEEYILHLLVLSHVYMVPDL 259
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
KRECEQKLELGLLT+DNLVDVFQLALLCDAPRLSLICHRKILKNF+ VS SEGWKAMKQS
Sbjct: 260 KRECEQKLELGLLTIDNLVDVFQLALLCDAPRLSLICHRKILKNFKAVSVSEGWKAMKQS 439
Query: 216 HPVLEKEILESMIDEENSKKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDF 275
HP+LEKEILES+IDEENSKKERIRK+NERE+YLQLYDAMEALVHICRDGCRTIGPHDKDF
Sbjct: 440 HPILEKEILESVIDEENSKKERIRKINEREIYLQLYDAMEALVHICRDGCRTIGPHDKDF 619
Query: 276 KANQPCRYTSCKGLELLVRHFAGCK 300
KANQPCRY +CK LELLVRHFAGCK
Sbjct: 620 KANQPCRYAACKALELLVRHFAGCK 694
Score = 26.6 bits (57), Expect(2) = e-108
Identities = 12/16 (75%), Positives = 12/16 (75%)
Frame = +1
Query: 78 LH*Y**RWHYLCSF*H 93
LH*Y* WH LCS *H
Sbjct: 1 LH*Y*KWWHCLCSL*H 48
>TC18112 similar to UP|Q9FJX5 (Q9FJX5) Genomic DNA, chromosome 5, TAC
clone:K9I9 (AT5g67480/K9I9_4), partial (18%)
Length = 469
Score = 102 bits (255), Expect = 7e-23
Identities = 44/45 (97%), Positives = 44/45 (97%)
Frame = +2
Query: 291 LLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
LLVRHFAGCKLRVPGGC HCKRMWQLLELHSRLCADPDFCRVPLC
Sbjct: 2 LLVRHFAGCKLRVPGGCVHCKRMWQLLELHSRLCADPDFCRVPLC 136
>TC14664 weakly similar to AAR24650 (AAR24650) At5g63160, partial (24%)
Length = 589
Score = 93.2 bits (230), Expect = 5e-20
Identities = 48/96 (50%), Positives = 63/96 (65%)
Frame = +2
Query: 107 RSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQKLELG 166
R I I GVP DAV F+ +LYS ++EM + +HLL LSHVY VPHLK+ C + L
Sbjct: 305 RIIQIHGVPGDAVTAFLTFLYSRRCTEDEMDRYGMHLLALSHVYMVPHLKQRCTKGLSQR 484
Query: 167 LLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRT 202
+ T +N+VD+ QLA LCDAP L L C + +NF+T
Sbjct: 485 VNT-ENVVDMLQLARLCDAPDLHLRCMNHLTRNFKT 589
>TC14373 weakly similar to UP|Q9FJX5 (Q9FJX5) Genomic DNA, chromosome 5, TAC
clone:K9I9 (AT5g67480/K9I9_4), partial (12%)
Length = 974
Score = 90.1 bits (222), Expect = 5e-19
Identities = 39/81 (48%), Positives = 53/81 (65%), Gaps = 2/81 (2%)
Frame = +2
Query: 257 LVHICRDGCRTIGPHDKDF-KANQPC-RYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMW 314
L HIC +GC +GPH + + + PC ++ +C+G++ L+RH+A C R+ GGC CKRMW
Sbjct: 2 LDHICTEGCTLVGPHHVEVDRKSGPCNKFATCQGVQQLIRHYATCTKRMGGGCLRCKRMW 181
Query: 315 QLLELHSRLCADPDFCRVPLC 335
QL LHS C D C VPLC
Sbjct: 182 QLFRLHSYGCEHADSCNVPLC 244
>TC17708 similar to UP|Q9FJX5 (Q9FJX5) Genomic DNA, chromosome 5, TAC
clone:K9I9 (AT5g67480/K9I9_4), partial (10%)
Length = 556
Score = 73.6 bits (179), Expect = 4e-14
Identities = 30/34 (88%), Positives = 31/34 (90%)
Frame = +2
Query: 302 RVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
RVPG C HC+RMW LLELHSRLCADPDFCRVPLC
Sbjct: 2 RVPGACVHCQRMWLLLELHSRLCADPDFCRVPLC 103
>TC11272 similar to UP|Q8LIQ8 (Q8LIQ8) OJ1370_E02.2 protein (OJ1354_H07.12
protein), partial (59%)
Length = 1070
Score = 40.8 bits (94), Expect = 3e-04
Identities = 34/138 (24%), Positives = 66/138 (47%), Gaps = 2/138 (1%)
Frame = +2
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEM-KEFVLHLLVLSHVYAVPH 154
ML+ +R +I I + +D + F+ YLY++ + + +LL L+ Y V H
Sbjct: 389 MLENDMEESRSGTIRITDISYDTLCSFVNYLYTAEAIASSLDSQMACNLLALAEKYQVNH 568
Query: 155 LKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILK-NFRTVSESEGWKAMK 213
LK CE+ L + L + + ++ A +A +L I+ N ++ E ++ +
Sbjct: 569 LKTFCEKYL-ISKLNWEKALALYSFANQHNANQLQDAALEFIINDNMAGLAAHEDYQDLV 745
Query: 214 QSHPVLEKEILESMIDEE 231
S+P L EI E+ + ++
Sbjct: 746 DSNPRLVVEIYEAYLTKQ 799
>BP047608
Length = 565
Score = 31.6 bits (70), Expect = 0.20
Identities = 18/51 (35%), Positives = 26/51 (50%)
Frame = +2
Query: 142 HLLVLSHVYAVPHLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLIC 192
HL+ + +Y + LK CE KL +T+DN+ LA P+L IC
Sbjct: 407 HLVAAADLYNIDRLKLLCESKL-CDEITIDNVATSLALAEQHHCPQLKAIC 556
>TC9929 similar to UP|O81396 (O81396) Aconitase-iron regulated protein 1,
partial (10%)
Length = 583
Score = 28.1 bits (61), Expect = 2.2
Identities = 18/42 (42%), Positives = 23/42 (53%)
Frame = +1
Query: 108 SISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHV 149
S SIF P+D V + L +SCY E M HL++ SHV
Sbjct: 409 SDSIFLFPYDVV--WRTTLLNSCYFLEVMLTMG*HLIIFSHV 528
>TC8213
Length = 935
Score = 27.7 bits (60), Expect = 2.8
Identities = 19/64 (29%), Positives = 26/64 (39%)
Frame = +2
Query: 136 MKEFVLHLLVLSHVYAVPHLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRK 195
M VL LL S ++ P C Q ++ LLTM N + + RL + H
Sbjct: 8 MSHIVLFLLGFSSLHGYP----TCSQGVKRSLLTMLNTLSIGSFLYWAGLTRLPYVHHLN 175
Query: 196 ILKN 199
L N
Sbjct: 176 CLSN 187
>TC17177 homologue to UP|GAPN_PEA (P81406) NADP-dependent
glyceraldehyde-3-phosphate dehydrogenase
(Non-phosphorylating glyceraldehyde 3-phosphate
dehydrogenase) (Glyceraldehyde-3-phosphate dehydrogenase
[NADP+]) (Triosephosphate dehydrogenase) , partial (40%)
Length = 689
Score = 26.9 bits (58), Expect = 4.8
Identities = 11/26 (42%), Positives = 16/26 (61%)
Frame = +1
Query: 262 RDGCRTIGPHDKDFKANQPCRYTSCK 287
R CR +G +++ KA+Q C Y CK
Sbjct: 346 RSNCRVLG--ERNSKASQRCSYRGCK 417
>TC8290
Length = 467
Score = 26.6 bits (57), Expect = 6.3
Identities = 11/29 (37%), Positives = 16/29 (54%), Gaps = 1/29 (3%)
Frame = +2
Query: 259 HICRDGCRTIGPHDKDFKANQPCR-YTSC 286
H C++ C GP F+ N PCR + +C
Sbjct: 380 HFCKNQCN*CGPFMNLFQ*NYPCRVFVTC 466
>TC14673 similar to UP|Q9LH76 (Q9LH76) Genomic DNA, chromosome 3, BAC clone:
T21E2 (AT3g14790/T21E2_4), partial (32%)
Length = 1048
Score = 26.2 bits (56), Expect = 8.2
Identities = 10/16 (62%), Positives = 12/16 (74%)
Frame = -3
Query: 320 HSRLCADPDFCRVPLC 335
HSR+C PD +VPLC
Sbjct: 434 HSRVCEVPDSPQVPLC 387
>TC18867 similar to UP|Q9ASV2 (Q9ASV2) At1g06730/F4H5_22, partial (25%)
Length = 748
Score = 26.2 bits (56), Expect = 8.2
Identities = 21/67 (31%), Positives = 30/67 (44%)
Frame = -1
Query: 102 RSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQ 161
R + SI +F H + R + + YSS + + HL SH + PH + C Q
Sbjct: 676 RDQQEPSIEVFHKRH*SYRQY-HHQYSS--QPYQCPHHEAHLTETSHRFHYPHDQLICSQ 506
Query: 162 KLELGLL 168
L L LL
Sbjct: 505 VLALRLL 485
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.342 0.149 0.512
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,147,064
Number of Sequences: 28460
Number of extensions: 115676
Number of successful extensions: 802
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 795
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 800
length of query: 335
length of database: 4,897,600
effective HSP length: 91
effective length of query: 244
effective length of database: 2,307,740
effective search space: 563088560
effective search space used: 563088560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146856.6


