

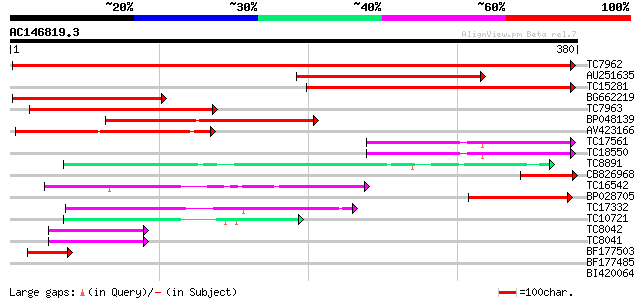
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146819.3 + phase: 0
(380 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC7962 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 ... 468 e-132
AU251635 242 7e-65
TC15281 similar to UP|Q8LJR2 (Q8LJR2) Alcohol dehydrogenase 1 (F... 209 6e-55
BG662219 204 2e-53
TC7963 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 ... 165 1e-41
BP048139 150 3e-37
AV423166 136 7e-33
TC17561 96 1e-20
TC18550 92 1e-19
TC8891 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Ara... 75 2e-14
CB826968 73 9e-14
TC16542 similar to UP|Q93X81 (Q93X81) Sorbitol dehydrogenase, pa... 71 3e-13
BP028705 64 3e-11
TC17332 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehyd... 59 1e-09
TC10721 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehyd... 54 4e-08
TC8042 homologue to UP|P93619 (P93619) Probable NADP-dependent o... 45 3e-05
TC8041 similar to UP|P93619 (P93619) Probable NADP-dependent oxi... 42 2e-04
BF177503 40 8e-04
BF177485 39 0.001
BI420064 35 0.021
>TC7962 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 , complete
Length = 1489
Score = 468 bits (1203), Expect = e-132
Identities = 215/377 (57%), Positives = 285/377 (75%)
Frame = +1
Query: 3 SSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPE 62
S+T GQVI C+AAV+WE KPL IE+VEVAPPQA EVR++IL+T+LCHTD Y W K
Sbjct: 115 STTAGQVIKCRAAVSWEAGKPLVIEEVEVAPPQAGEVRLKILYTSLCHTDVYFWEAKGQT 294
Query: 63 GLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAAT 122
LFP I GHEA GIVESVGEGVT +KPGDH +P + ECGEC CKS ++N+C +R T
Sbjct: 295 PLFPRIFGHEAGGIVESVGEGVTHLKPGDHALPVFTGECGECPHCKSEESNMCDLLRINT 474
Query: 123 GVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVP 182
GVM++D + RFSIKGKPIYHF+GTSTFS+YTV+H VAKI+P A LDKVC+L CG+
Sbjct: 475 DRGVMISDNQSRFSIKGKPIYHFVGTSTFSEYTVLHAGCVAKINPAAPLDKVCILSCGIC 654
Query: 183 TGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGV 242
TG GA N AK +PGS VAIFGLG VGLA AEGA+ +GASRIIG+D+ S++F+ AK FGV
Sbjct: 655 TGFGATVNVAKPKPGSSVAIFGLGAVGLAAAEGARVSGASRIIGVDLVSSRFEGAKKFGV 834
Query: 243 TEFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAAS 302
EF+NPKDH+KP+Q+VI+++T+GGVD + EC G++ M SA EC H GWG +V+VGV
Sbjct: 835 NEFVNPKDHDKPVQEVIAEMTNGGVDRAVECTGSIQAMISAFECVHDGWGVAVLVGVPNQ 1014
Query: 303 GQEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINE 362
T P + R KGT +G +K R+ +P +VE Y++ E+++++++TH ++ +EIN+
Sbjct: 1015DDAFQTHPVNFLNERTLKGTFYGNYKPRTDLPNVVEMYMRGELELEKFITHTVSFSEINQ 1194
Query: 363 AFDLMHEGKCLRCVLAL 379
AFD M +G+ +RC++ +
Sbjct: 1195AFDYMLKGESIRCIIRM 1245
>AU251635
Length = 383
Score = 242 bits (618), Expect = 7e-65
Identities = 118/127 (92%), Positives = 124/127 (96%)
Frame = +3
Query: 193 KVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTEFINPKDHE 252
KVEPGSIVA+FGLGTVGLAVAEGAK+AGASRIIGIDIDSNKF+ AKNFGVTEFINPK+HE
Sbjct: 3 KVEPGSIVAVFGLGTVGLAVAEGAKAAGASRIIGIDIDSNKFERAKNFGVTEFINPKEHE 182
Query: 253 KPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEISTRPFQ 312
KPIQQVI DLTDGGVDYSFECIGNVSVMR+ALEC HKGWGTSV+VGVAASGQEISTRPFQ
Sbjct: 183 KPIQQVIVDLTDGGVDYSFECIGNVSVMRAALECCHKGWGTSVIVGVAASGQEISTRPFQ 362
Query: 313 LVTGRVW 319
LVTGRVW
Sbjct: 363 LVTGRVW 383
>TC15281 similar to UP|Q8LJR2 (Q8LJR2) Alcohol dehydrogenase 1 (Fragment),
partial (50%)
Length = 785
Score = 209 bits (532), Expect = 6e-55
Identities = 92/180 (51%), Positives = 134/180 (74%)
Frame = +1
Query: 200 VAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTEFINPKDHEKPIQQVI 259
VA+FGLG VGLA AEGA+ +GASRIIG+D+ + +F+ AK FGVT+F+NPKDH+KP+QQVI
Sbjct: 4 VAVFGLGAVGLAAAEGARLSGASRIIGVDLVAGRFEQAKKFGVTDFVNPKDHDKPVQQVI 183
Query: 260 SDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEISTRPFQLVTGRVW 319
++T+G VD + EC GN+ SA EC H GWG +V+VGV E T P + GR
Sbjct: 184 VEMTNG*VDRAIECTGNIQATISAFECLHDGWGVAVIVGVPTKDAEFKTHPIHFMEGRTL 363
Query: 320 KGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAFDLMHEGKCLRCVLAL 379
KGT +G F++RS +P LVEKY+ KE+++++++TH++ +EIN+AF M +G+ +RC++ +
Sbjct: 364 KGTFYGNFQTRSDIPSLVEKYMNKELELEKFITHSVPFSEINQAFAYMLKGEGIRCLIRM 543
>BG662219
Length = 366
Score = 204 bits (519), Expect = 2e-53
Identities = 91/103 (88%), Positives = 101/103 (97%)
Frame = +2
Query: 3 SSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPE 62
++T+GQVITCKAAVAWEPNKPLTIEDV+VAPPQA EVR++IL+TALCHTDAYTWSGKDPE
Sbjct: 56 ATTQGQVITCKAAVAWEPNKPLTIEDVQVAPPQAGEVRVKILYTALCHTDAYTWSGKDPE 235
Query: 63 GLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECK 105
GLFPCILGHEAAG+VESVGEGVT+V+PGDHVIPCYQAEC ECK
Sbjct: 236 GLFPCILGHEAAGVVESVGEGVTEVQPGDHVIPCYQAECRECK 364
>TC7963 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 , partial
(33%)
Length = 694
Score = 165 bits (417), Expect = 1e-41
Identities = 77/126 (61%), Positives = 94/126 (74%)
Frame = +3
Query: 14 AAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEA 73
AAV+WE KPL IE+VEVAPPQA EVR++IL+T+LCHTD Y W K LFP I GHEA
Sbjct: 315 AAVSWEAGKPLVIEEVEVAPPQAGEVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEA 494
Query: 74 AGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGVGVMMNDRKP 133
GIVESVGEGVT +KP DH +P + ECGE CKS ++N+C +R T GVM++D++
Sbjct: 495 GGIVESVGEGVTHLKPVDHALPVFTGECGELPHCKSEESNMCDLLRINTDRGVMISDKQS 674
Query: 134 RFSIKG 139
RFSIKG
Sbjct: 675 RFSIKG 692
>BP048139
Length = 567
Score = 150 bits (379), Expect = 3e-37
Identities = 78/144 (54%), Positives = 95/144 (65%), Gaps = 1/144 (0%)
Frame = -2
Query: 65 FPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGV 124
FP ILGHEA G+VESVG+ VT+V GD VIP Y +CGEC CKS K+N C
Sbjct: 431 FPRILGHEAVGVVESVGKDVTEVTKGDVVIPIYLPDCGECTDCKSTKSNRCTNFPFKVSP 252
Query: 125 GVMMNDRKPRFS-IKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPT 183
M D RF+ + G+ I+HFM S+FS+YTVV +V KI P+ D+ CLL CG+ T
Sbjct: 251 S-MPRDGTTRFTDLNGEIIHHFMFISSFSEYTVVDVANVTKIDPEIPPDRACLLSCGIST 75
Query: 184 GLGAVWNTAKVEPGSIVAIFGLGT 207
G+GA W TA VEPGS VAIFGLG+
Sbjct: 74 GVGAAWRTASVEPGSTVAIFGLGS 3
>AV423166
Length = 452
Score = 136 bits (342), Expect = 7e-33
Identities = 67/134 (50%), Positives = 92/134 (68%)
Frame = +2
Query: 5 TEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGL 64
++ QVITCKAA+ W +P+T+E+++V PP+A EVR+++L ++CHTD + G P G+
Sbjct: 59 SKSQVITCKAAICWGLGEPVTVEEIQVDPPKATEVRVKMLCASICHTDITSIHGF-PHGI 235
Query: 65 FPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGV 124
FP LGHE GIVESVG+ VT++K GD +IP Y EC EC+ C SG+TNLC K A +
Sbjct: 236 FPLALGHEGVGIVESVGDQVTNLKEGDMIIPTYIGECQECENCVSGETNLCMKYPIA--L 409
Query: 125 GVMMNDRKPRFSIK 138
MM D R SI+
Sbjct: 410 SGMMPDNTSRMSIR 451
>TC17561
Length = 581
Score = 95.9 bits (237), Expect = 1e-20
Identities = 49/144 (34%), Positives = 85/144 (59%), Gaps = 4/144 (2%)
Frame = +3
Query: 240 FGVTEFINPKDHEKPIQQVISDLTDG-GVDYSFECIGNVSVMRSALECTHKGWGTSVVVG 298
FG+T FINP D K + +++ +L+ G GVDYS EC G ++ ++ T G G ++ VG
Sbjct: 6 FGMTHFINPGDSGKSLSELVKELSGGVGVDYSIECTGAPPLLTESVAATKVGTGKTIAVG 185
Query: 299 VAASGQEISTRPFQLVT---GRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNL 355
+A PF L+ GR KG+AFGG K+ + + + K LK+E + E TH +
Sbjct: 186 MAPE----PVVPFGLLALLFGRTLKGSAFGGIKTTTDLSIIANKCLKQEFPLQELFTHEV 353
Query: 356 TLAEINEAFDLMHEGKCLRCVLAL 379
L+++++AF+L+ + C++ ++ +
Sbjct: 354 PLSDVDKAFELLKQPDCVKIIIKM 425
>TC18550
Length = 688
Score = 92.0 bits (227), Expect = 1e-19
Identities = 50/144 (34%), Positives = 81/144 (55%), Gaps = 4/144 (2%)
Frame = +2
Query: 240 FGVTEFINPKDHEKPIQQVISDLTDG-GVDYSFECIGNVSVMRSALECTHKGWGTSVVVG 298
FG+T INP D K +++ +L+ G G+DYSFEC G + +LE T G G ++ +G
Sbjct: 14 FGMTHSINPGDSTKSASELVKELSGGIGMDYSFECSGFAPLPTESLEATKVGTGETIAIG 193
Query: 299 VAASGQEISTRPFQLVT---GRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNL 355
V PF +++ GR KG+ GG K+ S + K K+E + E TH +
Sbjct: 194 VGTE----PIVPFAILSILYGRTLKGSILGGLKAISDFSIIANKCQKEEFPLQELFTHEV 361
Query: 356 TLAEINEAFDLMHEGKCLRCVLAL 379
TLA+I++AF+L+ + C++ V+ +
Sbjct: 362 TLADISKAFELLIQSSCVKVVIKM 433
>TC8891 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Arabidopsis
thaliana;} , complete
Length = 1636
Score = 75.1 bits (183), Expect = 2e-14
Identities = 83/337 (24%), Positives = 128/337 (37%), Gaps = 8/337 (2%)
Frame = +1
Query: 37 NEVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVGEGVTDVKPGDHV-IP 95
++V ++I +C+ D ++PC+ GHE AGIV VG V GDHV +
Sbjct: 196 DDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVG 375
Query: 96 CYQAECGECKFCKSGKTNLCGKVRAATGVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYT 155
Y C +C+FC G + C K T GV D + G +S++
Sbjct: 376 TYVNSCRDCEFCDDGLEHHCVKGSVFTFNGV---DHDGTITKGG-----------YSKFI 513
Query: 156 VVHDVSVAKIHPDAALDKVCLLGCGVPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEG 215
VVH+ I L L C T + +PG + + GLG +G +
Sbjct: 514 VVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKF 693
Query: 216 AKSAGASRIIGIDIDSNKFDTAKNFGVTEFINPKDHEKPIQQVISDLTDGGVD-----YS 270
K+ G + + S K + G F+ D E+ + ++ D VD +S
Sbjct: 694 GKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEE--MRALAKSLDFIVDTASGPHS 867
Query: 271 FECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEISTRPFQLVTG-RVWKGTAFGGFKS 329
F+ K +G +VG EI P L+ G R G+ GG K
Sbjct: 868 FD----------PYMALLKSFGVLALVGFPG---EIKIHPGLLIMGSRTVSGSGVGGTKE 1008
Query: 330 -RSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAFD 365
R + + V + I+V + + NEA D
Sbjct: 1009IRDMIEFCVANEIHPNIEV-------VPIQYANEALD 1098
>CB826968
Length = 473
Score = 72.8 bits (177), Expect = 9e-14
Identities = 31/38 (81%), Positives = 36/38 (94%)
Frame = +1
Query: 343 KEIKVDEYVTHNLTLAEINEAFDLMHEGKCLRCVLALH 380
+EIKVDEY+THNLTL EIN+AFD+MHEG CLRCVLA+H
Sbjct: 271 QEIKVDEYITHNLTLVEINKAFDIMHEGGCLRCVLAMH 384
>TC16542 similar to UP|Q93X81 (Q93X81) Sorbitol dehydrogenase, partial (60%)
Length = 724
Score = 71.2 bits (173), Expect = 3e-13
Identities = 58/223 (26%), Positives = 94/223 (42%), Gaps = 5/223 (2%)
Frame = +1
Query: 24 LTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGLF---PCILGHEAAGIVESV 80
L I+ ++ ++VR+++ +C +D + P ++GHE AGI+E+V
Sbjct: 124 LKIQPFKLPTLGPHDVRVRMKAVGICGSDVHYLKKLRCADFIVKEPMVIGHECAGIIEAV 303
Query: 81 GEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGVGVMMNDRKPRFSIKGK 140
G V + PGD V C C CK G NLC P
Sbjct: 304 GAEVKSLVPGDRVAIEPGISCWRCDHCKLGSYNLC-----------------PDMKFFAT 432
Query: 141 PIYHFMGTSTFSQYTVVHDVSVA-KIHPDAALDKVCLLGCGVPTGLGA-VWNTAKVEPGS 198
P H S +Q +VH + K+ + +L++ + P +G A + P +
Sbjct: 433 PPVH---GSLANQ--IVHPADLCFKLPDNVSLEEGAMC---EPLSVGVHACRRANIGPET 588
Query: 199 IVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFG 241
V I G G +GL A++ GA RI+ +D+D ++ AK G
Sbjct: 589 NVLIMGAGPIGLVTMLAARAFGAPRIVIVDVDDHRLSVAKTLG 717
>BP028705
Length = 474
Score = 64.3 bits (155), Expect = 3e-11
Identities = 25/70 (35%), Positives = 49/70 (69%)
Frame = -1
Query: 308 TRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAFDLM 367
T P+ + G+ GTAFG +K ++ +P +V+ ++ ++++D+++TH + L+EIN+AFD M
Sbjct: 462 TSPYSFLDGKTLTGTAFGDYKPKTGLPSVVDLFMSNKLELDKFITHRVPLSEINKAFDYM 283
Query: 368 HEGKCLRCVL 377
+G LR ++
Sbjct: 282 IKGPSLRTII 253
>TC17332 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (63%)
Length = 736
Score = 58.9 bits (141), Expect = 1e-09
Identities = 50/200 (25%), Positives = 80/200 (40%), Gaps = 4/200 (2%)
Frame = +1
Query: 38 EVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVGEGVTDVKPGDHV-IPC 96
+V ++L+ +CH+D + + ++P + GHE AG+V VG V K GD V + C
Sbjct: 154 DVTFKVLYCGICHSDLHMIKNEWGSSVYPLVPGHEIAGVVTEVGSKVQKFKVGDRVGVGC 333
Query: 97 YQAECGECKFCKSGKTNLCGKVRAATGVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYT- 155
C C+ C N C K+ I + GT T+ Y+
Sbjct: 334 MVNSCRSCQSCADNLENYCPKM------------------ILTYAAKNVDGTITYGGYSD 459
Query: 156 -VVHDVSVA-KIHPDAALDKVCLLGCGVPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVA 213
+V D +I + L+ C T + +PG V + GLG +G
Sbjct: 460 LMVSDEDFGIRISDNLPLEAAAPFLCAGITVYSPLKYYGLDKPGLHVGVSGLGGLGHMAV 639
Query: 214 EGAKSAGASRIIGIDIDSNK 233
+ A + GA ++ I NK
Sbjct: 640 KFAIALGA-KVTAISTSPNK 696
>TC10721 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , complete
Length = 1297
Score = 53.9 bits (128), Expect = 4e-08
Identities = 45/169 (26%), Positives = 64/169 (37%), Gaps = 8/169 (4%)
Frame = +3
Query: 37 NEVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVGEGVTDVKPGDHV-IP 95
++V I+ILF +CH+D +T +P + GHE G+V G VT K GD V +
Sbjct: 186 DDVAIKILFCGVCHSDLHTIKNDWGFTTYPVVPGHEIVGVVTKTGNNVTKFKVGDKVGVG 365
Query: 96 CYQAECGECKFCKSGKTNLCGKVRAATGVGVMMNDRKPRFSIKGKPIY----HFMGTST- 150
C C+ C N C +P+Y + GT T
Sbjct: 366 VIVDSCKSCENCDQDLENYC-----------------------PQPVYTYNSPYNGTRTN 476
Query: 151 --FSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTGLGAVWNTAKVEPG 197
+S + VVH V + + LD L C T + EPG
Sbjct: 477 GGYSDFVVVHQRYVLQFPDNLPLDAGAPLLCAGITVYXPMKYYGMTEPG 623
>TC8042 homologue to UP|P93619 (P93619) Probable NADP-dependent
oxidoreductase TED2 , complete
Length = 1244
Score = 44.7 bits (104), Expect = 3e-05
Identities = 26/67 (38%), Positives = 34/67 (49%)
Frame = +2
Query: 27 EDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVGEGVTD 86
EDVE+ P+ NEVR++ + D Y G P G EA G+V +VG G T
Sbjct: 170 EDVEIGDPKENEVRVKNKAIGVNFIDVYFRKGVYKASTSPFTPGMEAVGVVTAVGGGPTG 349
Query: 87 VKPGDHV 93
V+ GD V
Sbjct: 350 VQVGDLV 370
>TC8041 similar to UP|P93619 (P93619) Probable NADP-dependent
oxidoreductase TED2 , partial (31%)
Length = 637
Score = 42.0 bits (97), Expect = 2e-04
Identities = 25/67 (37%), Positives = 33/67 (48%)
Frame = +1
Query: 27 EDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVGEGVTD 86
EDVE+ P+ EVR++ + D Y G P G EA G+V +VG G T
Sbjct: 394 EDVEIGEPKEGEVRVKNKAIGVNFIDVYFRKGVYKASSTPFTPGMEAVGVVTAVGGGPTG 573
Query: 87 VKPGDHV 93
V+ GD V
Sbjct: 574 VQVGDLV 594
>BF177503
Length = 355
Score = 39.7 bits (91), Expect = 8e-04
Identities = 17/30 (56%), Positives = 22/30 (72%)
Frame = +1
Query: 13 KAAVAWEPNKPLTIEDVEVAPPQANEVRIQ 42
+ AV WEPNKPLTIE+ + P+A EV I+
Sbjct: 223 RGAVFWEPNKPLTIEEFHMPRPEAGEVLIK 312
>BF177485
Length = 334
Score = 39.3 bits (90), Expect = 0.001
Identities = 22/54 (40%), Positives = 28/54 (51%)
Frame = +1
Query: 28 DVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVG 81
DV V P+ EV ++ F + + D Y SG FP ILG E AG + SVG
Sbjct: 136 DVPVPEPKEGEVLVKNEFIGINYIDTYFRSGVYKPPQFPYILGREGAGTIASVG 297
>BI420064
Length = 410
Score = 35.0 bits (79), Expect = 0.021
Identities = 24/70 (34%), Positives = 35/70 (49%), Gaps = 3/70 (4%)
Frame = +3
Query: 28 DVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGL---FPCILGHEAAGIVESVGEGV 84
+V + P +EV I++ ++ D GK L FP I G + AG V +G+GV
Sbjct: 168 EVPIPAPSKDEVLIKLEAASINPFDWKVQIGKLRPLLPRKFPYIPGTDIAGEVVEIGQGV 347
Query: 85 TDVKPGDHVI 94
KPGD V+
Sbjct: 348 KKFKPGDKVV 377
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,885,978
Number of Sequences: 28460
Number of extensions: 91875
Number of successful extensions: 412
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 403
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 404
length of query: 380
length of database: 4,897,600
effective HSP length: 92
effective length of query: 288
effective length of database: 2,279,280
effective search space: 656432640
effective search space used: 656432640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146819.3


