

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146817.8 + phase: 0
(831 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
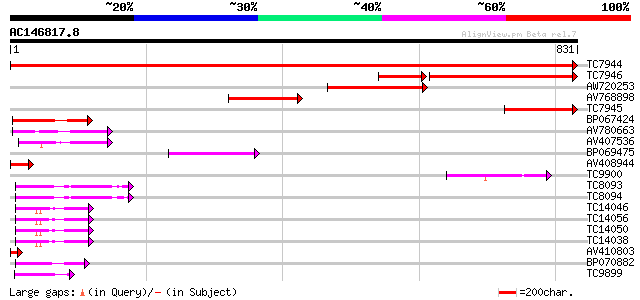
Score E
Sequences producing significant alignments: (bits) Value
TC7944 homologue to UP|Q9ASR1 (Q9ASR1) At1g56070/T6H22_13 (Elong... 1473 0.0
TC7946 homologue to UP|EF2_BETVU (O23755) Elongation factor 2 (E... 365 e-131
AW720253 204 4e-53
AV768898 199 1e-51
TC7945 homologue to UP|Q9ASR1 (Q9ASR1) At1g56070/T6H22_13 (Elong... 172 2e-43
BP067424 107 8e-24
AV780663 88 5e-18
AV407536 75 3e-14
BP069475 73 2e-13
AV408944 71 6e-13
TC9900 homologue to UP|EFGC_SOYBN (P34811) Elongation factor G, ... 60 2e-09
TC8093 homologue to UP|EFTU_TOBAC (P41342) Elongation factor Tu,... 58 5e-09
TC8094 homologue to UP|EFTU_PEA (O24310) Elongation factor Tu, c... 57 1e-08
TC14046 homologue to UP|P93769 (P93769) Elongation factor-1 alph... 50 1e-06
TC14056 homologue to UP|P93769 (P93769) Elongation factor-1 alph... 48 7e-06
TC14050 homologue to UP|EF1A_TOBAC (P43643) Elongation factor 1-... 48 7e-06
TC14038 homologue to UP|Q9LN13 (Q9LN13) T6D22.2, partial (47%) 48 7e-06
AV410803 44 1e-04
BP070882 44 1e-04
TC9899 homologue to PIR|S35701|S35701 translation elongation fac... 42 4e-04
>TC7944 homologue to UP|Q9ASR1 (Q9ASR1) At1g56070/T6H22_13 (Elongation factor
EF-2), complete
Length = 2971
Score = 1473 bits (3814), Expect = 0.0
Identities = 719/831 (86%), Positives = 771/831 (92%)
Frame = +1
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTR DEAERGITIKS
Sbjct: 244 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKS 423
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYYEM+D LK+FKGER GN+YLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV
Sbjct: 424 TGISLYYEMTDVALKSFKGERMGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 603
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLEL +D EEAY T RVIE+ NV+MATY
Sbjct: 604 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELQVDGEEAYQTFSRVIENANVIMATY 783
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
ED LLGD VYPEKGTV+FSAGLHGW+FTLTNFAKMYASKFGVDE KMM RLWGENFFD
Sbjct: 784 EDPLLGDCMVYPEKGTVAFSAGLHGWAFTLTNFAKMYASKFGVDESKMMERLWGENFFDP 963
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
+TKKWT+K+T + TCKRGFVQFCYEPIKQII CMNDQKDKLWPMLQKLGV +KSEEK+L
Sbjct: 964 ATKKWTSKNTGSATCKRGFVQFCYEPIKQIINTCMNDQKDKLWPMLQKLGVTMKSEEKDL 1143
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
GK LMKRVMQ+WLPA+SALLEMMIFHLPSP+ AQ+YRVENLYEGPLDD YA+AIRNCDP
Sbjct: 1144 MGKPLMKRVMQTWLPAASALLEMMIFHLPSPSTAQRYRVENLYEGPLDDQYAAAIRNCDP 1323
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
EGPLMLYVSKMIPASDKGRF+AFGRVFSGKVSTG+KVRIMGPNY+PGEKKDLY KSVQRT
Sbjct: 1324 EGPLMLYVSKMIPASDKGRFFAFGRVFSGKVSTGLKVRIMGPNYVPGEKKDLYTKSVQRT 1503
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGKKQETVEDVPCGNTVA+VGLDQFITKNATLTNEKE DAHPIRAMKFSVSPVV VA
Sbjct: 1504 VIWMGKKQETVEDVPCGNTVALVGLDQFITKNATLTNEKETDAHPIRAMKFSVSPVVRVA 1683
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
V CKVASDLPKLVEGLKRLAKSDPMVVCTI E+GEHI+A AGELHLEICLKDLQDDFM G
Sbjct: 1684 VQCKVASDLPKLVEGLKRLAKSDPMVVCTIEESGEHIVAGAGELHLEICLKDLQDDFMGG 1863
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEI KSDP+VSFRETVLE+S TVMSKSPNKHNRLYMEARP+E+GLAEAIDDG+IGPRD+
Sbjct: 1864 AEIVKSDPVVSFRETVLERSCRTVMSKSPNKHNRLYMEARPLEDGLAEAIDDGKIGPRDD 2043
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PK KILS+E+GWDKDLAKK+WCFGPET GPNM+VD CKGVQYLNEIKDSVVAGFQ AS
Sbjct: 2044 PKVRSKILSEEYGWDKDLAKKIWCFGPETLGPNMVVDMCKGVQYLNEIKDSVVAGFQWAS 2223
Query: 661 KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV 720
KEG +A+EN+R +CFEVCDVVLH DAIHRGGGQIIPTARRVFYA+ LTAKPRLLEPVYLV
Sbjct: 2224 KEGALAEENMRAICFEVCDVVLHADAIHRGGGQIIPTARRVFYASQLTAKPRLLEPVYLV 2403
Query: 721 EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
EIQAPE ALGGIYSVLNQKRGHVF+E+QR TPLYN+KAYLPV+ESF F+ +LRA T GQ
Sbjct: 2404 EIQAPEQALGGIYSVLNQKRGHVFEEMQRQGTPLYNIKAYLPVVESFGFSGTLRAATSGQ 2583
Query: 781 AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
AFPQ VFDHWDM+ SDPLE G+ AA V +IRK+KGLKEQ+ PLSEFED+L
Sbjct: 2584 AFPQCVFDHWDMMSSDPLESGSQAATLVTDIRKRKGLKEQMTPLSEFEDKL 2736
>TC7946 homologue to UP|EF2_BETVU (O23755) Elongation factor 2 (EF-2),
partial (34%)
Length = 1009
Score = 365 bits (936), Expect(2) = e-131
Identities = 173/216 (80%), Positives = 192/216 (88%)
Frame = +3
Query: 616 KDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIASKEGPMADENLRGVCF 675
K KK+WCFGPET GPNM+VD CKGVQYLNEIKDSVVAGFQ ASKEG +A+EN+R +CF
Sbjct: 198 KSCPKKIWCFGPETLGPNMVVDMCKGVQYLNEIKDSVVAGFQWASKEGALAEENMRAICF 377
Query: 676 EVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLVEIQAPEHALGGIYSV 735
EVCDVVLH DAIHRGGGQIIPTARRVFYA+ LTAKPRLLEPVYLVEIQAPE ALGGIYSV
Sbjct: 378 EVCDVVLHADAIHRGGGQIIPTARRVFYASQLTAKPRLLEPVYLVEIQAPEQALGGIYSV 557
Query: 736 LNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQAFPQLVFDHWDMVPS 795
LNQKRGHVF+E+QRP TPLYN+KAYLPV+ESF F+ +LRA T GQAFPQ VFDHWDM+ S
Sbjct: 558 LNQKRGHVFEEMQRPGTPLYNIKAYLPVVESFGFSGTLRAATSGQAFPQCVFDHWDMMSS 737
Query: 796 DPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
DPLE G+ AA V +IRK+KGLKEQ+ PLSEFED+L
Sbjct: 738 DPLESGSQAATLVTDIRKRKGLKEQMTPLSEFEDKL 845
Score = 121 bits (303), Expect(2) = e-131
Identities = 58/71 (81%), Positives = 65/71 (90%)
Frame = +2
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEI KSDP+VSFRETVLE+S TVMSKSPNKHNRLYMEARP+E+GLAEAIDDG+IGPRD+
Sbjct: 2 AEIVKSDPVVSFRETVLERSCRTVMSKSPNKHNRLYMEARPLEDGLAEAIDDGKIGPRDD 181
Query: 601 PKNHLKILSDE 611
PK KILS+E
Sbjct: 182 PKVRSKILSEE 214
>AW720253
Length = 481
Score = 204 bits (520), Expect = 4e-53
Identities = 99/146 (67%), Positives = 119/146 (80%)
Frame = +3
Query: 467 RAMKFSVSPVVSVAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHL 526
R KFSVSPVV VAV K +DLPKLVEGLKRL+KSDP V C + E+GEHI+A AGELHL
Sbjct: 42 RVGKFSVSPVVRVAVEPKNPADLPKLVEGLKRLSKSDPCVQCFMEESGEHIVAGAGELHL 221
Query: 527 EICLKDLQDDFMNGAEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGL 586
EICLKDLQDDFMNG E+ +DPIVS+RETV S H V+SKSPNKHNR+Y++A PM++GL
Sbjct: 222 EICLKDLQDDFMNGVELKITDPIVSYRETVTAPSDHVVLSKSPNKHNRIYLKAEPMQDGL 401
Query: 587 AEAIDDGRIGPRDEPKNHLKILSDEF 612
AEAI+ G++GPRD+PK K L++ F
Sbjct: 402 AEAIEGGKVGPRDDPKVRGKFLAENF 479
>AV768898
Length = 328
Score = 199 bits (506), Expect = 1e-51
Identities = 94/109 (86%), Positives = 101/109 (92%)
Frame = +2
Query: 321 LEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDPEGPLMLYVSKMIPASDKGRF 380
LEMMIF+LPSP+ AQ+YRVENLYEGPLDD YA+AIRNCDPEGPLMLYV KMIPASDKG F
Sbjct: 2 LEMMIFYLPSPSTAQRYRVENLYEGPLDDQYATAIRNCDPEGPLMLYVLKMIPASDKGMF 181
Query: 381 YAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRTVIWMGKKQE 429
+AFGRVFSGKVSTG+KV IMGPNY+PGEKKDLY KS RTVIWMGKKQE
Sbjct: 182 FAFGRVFSGKVSTGLKVTIMGPNYVPGEKKDLYTKSGHRTVIWMGKKQE 328
>TC7945 homologue to UP|Q9ASR1 (Q9ASR1) At1g56070/T6H22_13 (Elongation
factor EF-2), partial (13%)
Length = 712
Score = 172 bits (436), Expect = 2e-43
Identities = 80/106 (75%), Positives = 93/106 (87%)
Frame = +2
Query: 726 EHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQAFPQL 785
E ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPV+ESF F+ +LRA T GQAFPQ
Sbjct: 2 EQALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVVESFGFSSTLRAATSGQAFPQC 181
Query: 786 VFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
VFDHWDM+ SDPLEPG+ AA V +IR++KGLKEQ+ PLSEFED+L
Sbjct: 182 VFDHWDMMSSDPLEPGSGAAQLVADIRRRKGLKEQMTPLSEFEDKL 319
>BP067424
Length = 473
Score = 107 bits (267), Expect = 8e-24
Identities = 56/120 (46%), Positives = 77/120 (63%), Gaps = 2/120 (1%)
Frame = +1
Query: 4 KHNIRNMSVIAHVDHGKSTLTDSLVAAA--GIIAQEVAGDVRMTDTRQDEAERGITIKST 61
+H IRN+ ++AHVDHGK+TL D L+A A G++ +VAG VR D +E R IT+KS+
Sbjct: 160 RHKIRNICILAHVDHGKTTLADQLIAVASGGMVHPKVAGRVRFMDYLDEEQRRAITMKSS 339
Query: 62 GISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVE 121
I+L Y N + +NLIDSPGH+DF EV+ A R++DGAL++VD VE
Sbjct: 340 SIALRY----------------NHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVE 471
>AV780663
Length = 568
Score = 88.2 bits (217), Expect = 5e-18
Identities = 54/147 (36%), Positives = 82/147 (55%), Gaps = 1/147 (0%)
Frame = +1
Query: 5 HNIRNMSVIAHVDHGKSTLTDSLVAAAGI-IAQEVAGDVRMTDTRQDEAERGITIKSTGI 63
+ +RN++VIAHVDHGK+TL D ++ G I E R D+ E ERGITI S
Sbjct: 160 NRLRNVAVIAHVDHGKTTLMDRILRQCGADIPHE-----RAMDSNSLERERGITIASKVT 324
Query: 64 SLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGV 123
++ ++ ++ +N++D+PGH D EV + + +GA+ VVD EG
Sbjct: 325 TVSWKENE----------------LNMVDTPGHADLGGEVERVVGMVEGAV*VVDAGEGP 456
Query: 124 CVQTETVLRQALGERIKPVLTVNKMDR 150
QT+ VL +AL ++P+L NK+DR
Sbjct: 457 LAQTKLVLAKALKYGLRPLLLKNKVDR 537
>AV407536
Length = 382
Score = 75.5 bits (184), Expect = 3e-14
Identities = 55/142 (38%), Positives = 79/142 (54%), Gaps = 5/142 (3%)
Frame = +2
Query: 14 AHVDHGKSTLTDSLVAAAGIIAQ--EVAGDVRM---TDTRQDEAERGITIKSTGISLYYE 68
AH+D GK+TLT+ ++ AG I + EV G + D+ E E+GITI+S ++
Sbjct: 5 AHIDSGKTTLTERVLFYAGRIHEIHEVRGRDGVGAKMDSMDLEREKGITIQSAATYCSWK 184
Query: 69 MSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQTE 128
D K IN+ID+PGHVDF+ EV ALR+ DGA++V+ V GV Q+
Sbjct: 185 ----DCK------------INIIDTPGHVDFTIEVERALRVLDGAILVLCSVGGVQSQSI 316
Query: 129 TVLRQALGERIKPVLTVNKMDR 150
TV RQ + + +NK+DR
Sbjct: 317 TVDRQMRRYDVPRLAFINKLDR 382
>BP069475
Length = 442
Score = 72.8 bits (177), Expect = 2e-13
Identities = 39/134 (29%), Positives = 68/134 (50%)
Frame = +2
Query: 233 WGENFFDSSTKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVN 292
WG+ +F T+ + K + +R FV+F EP+ +I + + K + L +LGV
Sbjct: 44 WGDYYFHPDTRTFKKKPPVSGG-ERSFVEFVLEPLYKIYSQVIGEHKKSVETTLAELGVT 220
Query: 293 LKSEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYA 352
L + L+ + L++ S +S +M++ H+PSP A +V+++Y G D
Sbjct: 221 LSNAAYRLNVRPLLRLACSSVFGQASGFTDMLVQHIPSPRDAAVKKVDHIYTGLKDSSIY 400
Query: 353 SAIRNCDPEGPLML 366
A+ CD GPLM+
Sbjct: 401 KAMTQCDSSGPLMV 442
>AV408944
Length = 295
Score = 71.2 bits (173), Expect = 6e-13
Identities = 35/35 (100%), Positives = 35/35 (100%)
Frame = +1
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIA 35
MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIA
Sbjct: 190 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIA 294
>TC9900 homologue to UP|EFGC_SOYBN (P34811) Elongation factor G,
chloroplast precursor (EF-G), partial (34%)
Length = 1284
Score = 59.7 bits (143), Expect = 2e-09
Identities = 49/161 (30%), Positives = 74/161 (45%), Gaps = 7/161 (4%)
Frame = +1
Query: 641 GVQYLNEIKDSVVAGFQIASKEGPMADENLRGVC--FEVCDV--VLHTDAIHRGGGQIIP 696
G ++ +EIK V I + + GV F V DV VL + H ++
Sbjct: 436 GYEFKSEIKGGAVPREYIPGVMKGLEECMSNGVLAGFPVVDVRAVLTDGSYHDVDSSVLA 615
Query: 697 ---TARRVFYAAMLTAKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTP 753
AR F + A PR+LEP+ VE+ PE LG + LN +RG + +P
Sbjct: 616 FQLAARGAFREGVRKAGPRMLEPIMKVEVVTPEEHLGDVIGDLNSRRGQINSFGDKPG-G 792
Query: 754 LYNVKAYLPVIESFQFNESLRAQTGGQAFPQLVFDHWDMVP 794
L V + +P+ E FQ+ +LR T G+A + +D+VP
Sbjct: 793 LKVVDSLVPLAEMFQYVSTLRGMTKGRASYTMQLAMFDVVP 915
>TC8093 homologue to UP|EFTU_TOBAC (P41342) Elongation factor Tu,
chloroplast precursor (EF-Tu), partial (58%)
Length = 1146
Score = 58.2 bits (139), Expect = 5e-09
Identities = 50/175 (28%), Positives = 76/175 (42%), Gaps = 2/175 (1%)
Frame = +1
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLYYE 68
N+ I HVDHGK+TLT +L A + D +E RGITI + +
Sbjct: 358 NIGTIGHVDHGKTTLTAALTMALASLGNSAPKKYDEIDAAPEERARGITINTATVEY--- 528
Query: 69 MSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQTE 128
E E Y +D PGH D+ + DGA++VV +G QT+
Sbjct: 529 -----------ETENRHYA--HVDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQTK 669
Query: 129 --TVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATYE 181
+L + +G V+ +NK D+ +D EE +Q V V ++++YE
Sbjct: 670 EHILLAKQVGVP-NMVVFLNKQDQ------VDDEE---LLQLVELEVRELLSSYE 804
>TC8094 homologue to UP|EFTU_PEA (O24310) Elongation factor Tu, chloroplast
precursor (EF-Tu), partial (60%)
Length = 1092
Score = 57.0 bits (136), Expect = 1e-08
Identities = 47/174 (27%), Positives = 73/174 (41%), Gaps = 1/174 (0%)
Frame = +1
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLYYE 68
N+ I HVDHGK+TLT +L A + D +E RGITI + +
Sbjct: 331 NIGTIGHVDHGKTTLTAALTMALASLGNSAPKKYDEIDAAPEERARGITINTATVEY--- 501
Query: 69 MSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQTE 128
E E Y +D PGH D+ + DGA++VV +G QT+
Sbjct: 502 -----------ETETRHYA--HVDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQTK 642
Query: 129 TVLRQALGERIKPVLT-VNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATYE 181
+ A + V+ +NK D+ +D EE ++ V ++++YE
Sbjct: 643 EHILLAKQVGVPNVVVFLNKQDQ------VDDEELLELVEL---EVRELLSSYE 777
>TC14046 homologue to UP|P93769 (P93769) Elongation factor-1 alpha, partial
(84%)
Length = 1506
Score = 50.1 bits (118), Expect = 1e-06
Identities = 41/129 (31%), Positives = 58/129 (44%), Gaps = 15/129 (11%)
Frame = +1
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEV-------AGDVR--------MTDTRQDEAE 53
N+ VI HVD GKST T L+ G I + V A ++ + D + E E
Sbjct: 106 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 285
Query: 54 RGITIKSTGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGA 113
RGITI I+L+ + E NKY +ID+PGH DF + D A
Sbjct: 286 RGITID---IALW-------------KFETNKYYCTVIDAPGHRDFIKNMITGTSQADCA 417
Query: 114 LVVVDCVEG 122
++++D G
Sbjct: 418 VLIIDSTTG 444
>TC14056 homologue to UP|P93769 (P93769) Elongation factor-1 alpha, complete
Length = 1897
Score = 47.8 bits (112), Expect = 7e-06
Identities = 40/129 (31%), Positives = 57/129 (44%), Gaps = 15/129 (11%)
Frame = +1
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEV-------AGDVR--------MTDTRQDEAE 53
N+ VI HVD GKST T L+ G I + V A ++ + D + E E
Sbjct: 133 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 312
Query: 54 RGITIKSTGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGA 113
RGITI I+L+ + E KY +ID+PGH DF + D A
Sbjct: 313 RGITI---DIALW-------------KFETTKYYCTVIDAPGHRDFIKNMITGTSQADCA 444
Query: 114 LVVVDCVEG 122
++++D G
Sbjct: 445 VLIIDSTTG 471
>TC14050 homologue to UP|EF1A_TOBAC (P43643) Elongation factor 1-alpha
(EF-1-alpha) (Vitronectin-like adhesion protein 1)
(PVN1), partial (38%)
Length = 583
Score = 47.8 bits (112), Expect = 7e-06
Identities = 40/129 (31%), Positives = 57/129 (44%), Gaps = 15/129 (11%)
Frame = +2
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEV-------AGDVR--------MTDTRQDEAE 53
N+ VI HVD GKST T L+ G I + V A ++ + D + E E
Sbjct: 95 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 274
Query: 54 RGITIKSTGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGA 113
RGITI I+L+ + E KY +ID+PGH DF + D A
Sbjct: 275 RGITI---DIALW-------------KFETTKYYCTVIDAPGHRDFIKNMITGTSQADCA 406
Query: 114 LVVVDCVEG 122
++++D G
Sbjct: 407 VLIIDSTTG 433
>TC14038 homologue to UP|Q9LN13 (Q9LN13) T6D22.2, partial (47%)
Length = 1844
Score = 47.8 bits (112), Expect = 7e-06
Identities = 40/129 (31%), Positives = 57/129 (44%), Gaps = 15/129 (11%)
Frame = +3
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEV-------AGDVR--------MTDTRQDEAE 53
N+ VI HVD GKST T L+ G I + V A ++ + D + E E
Sbjct: 114 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 293
Query: 54 RGITIKSTGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGA 113
RGITI I+L+ + E KY +ID+PGH DF + D A
Sbjct: 294 RGITI---DIALW-------------KFETTKYYCTVIDAPGHRDFIKNMITGTSQADCA 425
Query: 114 LVVVDCVEG 122
++++D G
Sbjct: 426 VLIIDSTTG 452
>AV410803
Length = 135
Score = 43.9 bits (102), Expect = 1e-04
Identities = 19/19 (100%), Positives = 19/19 (100%)
Frame = +3
Query: 1 MDLKHNIRNMSVIAHVDHG 19
MDLKHNIRNMSVIAHVDHG
Sbjct: 78 MDLKHNIRNMSVIAHVDHG 134
>BP070882
Length = 496
Score = 43.5 bits (101), Expect = 1e-04
Identities = 32/109 (29%), Positives = 48/109 (43%), Gaps = 1/109 (0%)
Frame = +2
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLYYE 68
N+ + HVDHGK+TLT ++ + A D +E +RGITI + +
Sbjct: 209 NVGTMGHVDHGKTTLTAAITKVLADEGKAKAIAFDEIDKAPEEKKRGITIATAHV----- 373
Query: 69 MSDGDLKNFKGEREGNKYLINLIDSPGHVDF-SSEVTAALRITDGALVV 116
E E K +D PGH D+ + +T A ++ G LVV
Sbjct: 374 -----------EYETVKRHYAHVDCPGHADYVKNMITGAAQMDGGILVV 487
>TC9899 homologue to PIR|S35701|S35701 translation elongation factor EF-G,
chloroplast - soybean {Glycine max;} , partial (15%)
Length = 624
Score = 42.0 bits (97), Expect = 4e-04
Identities = 27/89 (30%), Positives = 44/89 (49%), Gaps = 2/89 (2%)
Frame = +1
Query: 8 RNMSVIAHVDHGKSTLTDSLVAAAGIIAQ--EVAGDVRMTDTRQDEAERGITIKSTGISL 65
RN+ ++AH+D GK+T T+ ++ G + EV D + E ERGITI S +
Sbjct: 406 RNIGIMAHIDAGKTTTTERILFYTGRNYKIGEVHEGTATMDWMEQEQERGITITSAATTT 585
Query: 66 YYEMSDGDLKNFKGEREGNKYLINLIDSP 94
++ NK+ I++ID+P
Sbjct: 586 FW----------------NKHRIHIIDTP 624
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,536,600
Number of Sequences: 28460
Number of extensions: 156871
Number of successful extensions: 734
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 725
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 726
length of query: 831
length of database: 4,897,600
effective HSP length: 98
effective length of query: 733
effective length of database: 2,108,520
effective search space: 1545545160
effective search space used: 1545545160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146817.8


