

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146805.1 - phase: 0 /pseudo
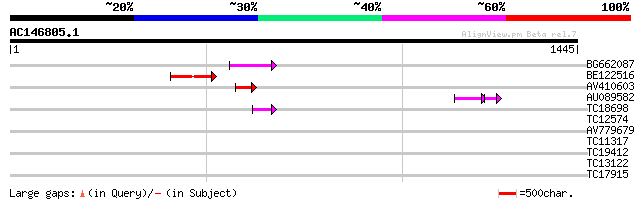
(1445 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG662087 91 1e-18
BE122516 85 7e-17
AV410603 72 8e-13
AU089582 50 1e-08
TC18698 44 2e-04
TC12574 37 0.017
AV779679 34 0.19
TC11317 similar to UP|Q40363 (Q40363) NuM1 protein, partial (12%) 30 3.5
TC19412 similar to UP|Q84KB0 (Q84KB0) Pol protein, partial (7%) 29 4.6
TC13122 similar to UP|Q84Y01 (Q84Y01) Inositol phosphate kinase,... 29 4.6
TC17915 similar to GB|AAM10415.1|20147411|AY093799 AT3g05640/F18... 28 7.9
>BG662087
Length = 373
Score = 90.9 bits (224), Expect = 1e-18
Identities = 45/119 (37%), Positives = 67/119 (55%)
Frame = +1
Query: 560 GSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGARVFSKIDLRSGYHQIKVKDEDMQK 619
G R+ +DY LNK K+ YPLP ID L+D + S +D SGYHQIK+ D K
Sbjct: 16 GKWRMWVDYTDLNKACPKDSYPLPSIDKLVDGASDNELLSLMDAYSGYHQIKMHPSDEDK 195
Query: 620 TALRTRYGHYEYKVMPFGVTNAPRVFMEYMNRIFHAFLDRFVVVFIDDILIYSKTEEEH 678
TA T +Y Y+ +PFG+ NA + M+R+F + R + V++D++++ S H
Sbjct: 196 TAFMTARVNYCYQTIPFGLKNAGATYQXLMDRVFXDXVGRNMEVYLDNMIVKSALRANH 372
>BE122516
Length = 364
Score = 85.1 bits (209), Expect = 7e-17
Identities = 44/116 (37%), Positives = 72/116 (61%)
Frame = +2
Query: 411 LGMNWLEYKHVHINCFSKSVYFSSAEEECGAEFLSTKQLKQMERDGILMFSLMASLSIEN 470
+GMNWL +NC K+V F ++E + + K K E + ++ + + ++
Sbjct: 14 VGMNWLTANDATLNCRKKTVTFGTSEGDAKRVKRTDKVGKASECESDVLLGALET--DKS 187
Query: 471 QAVIDKLQVVCDFPEVYPDEIPDVPPEREVEFSINLVPGTKPVSMAPYRMSASELS 526
++ + VV +F +V+P+E+ ++PPEREVEFSI+ VPGT P+S+APYRMS EL+
Sbjct: 188 DTGVEGIPVVREFSDVFPEEVSELPPEREVEFSID*VPGTGPISIAPYRMSLVELA 355
>AV410603
Length = 162
Score = 71.6 bits (174), Expect = 8e-13
Identities = 30/53 (56%), Positives = 42/53 (78%)
Frame = +1
Query: 575 TIKNRYPLPRIDDLMDQLVGARVFSKIDLRSGYHQIKVKDEDMQKTALRTRYG 627
T+K+ +P+P +D+L+D+L G++ FSK+DLRSGYHQI VK ED KT RT +G
Sbjct: 4 TVKDSFPMPTVDELLDELRGSQFFSKLDLRSGYHQILVKPEDRHKTVFRTHHG 162
>AU089582
Length = 383
Score = 50.1 bits (118), Expect(2) = 1e-08
Identities = 30/83 (36%), Positives = 42/83 (50%)
Frame = +1
Query: 1133 CCPVGGDLH*GDCEVAWYSFEHCIG*RSKIYF*ILEKFARGFGFEVEIEFGLSSTDRWSV 1192
C + DL DC +AW + + RS I+ LE F+ FG ++ E+ SS++ WSV
Sbjct: 10 CVSIC*DLLG*DCFLAWCTCVYNFRSRSSIHITFLEVFSNCFGNSIKNEYRFSSSN*WSV 189
Query: 1193 GEDNSVARGFVESLCA*ARRNLG 1215
ED S RG+ LC R G
Sbjct: 190 REDYSDLRGYASCLCVXPERXAG 258
Score = 27.3 bits (59), Expect(2) = 1e-08
Identities = 20/43 (46%), Positives = 24/43 (55%)
Frame = +2
Query: 1211 RRNLG*SSSVDRVHIQ**LSF*YWNGTF*GFVWSEMQNSVVLV 1253
R LG D + I **LS *+ +GT * VW EMQ + LV
Sbjct: 245 RGXLGSVFVFDGIRI***LSV*HLDGTI*SLVW*EMQVTYRLV 373
>TC18698
Length = 808
Score = 43.5 bits (101), Expect = 2e-04
Identities = 20/61 (32%), Positives = 36/61 (58%)
Frame = -2
Query: 618 QKTALRTRYGHYEYKVMPFGVTNAPRVFMEYMNRIFHAFLDRFVVVFIDDILIYSKTEEE 677
+KT L+ +Y Y+VMP G+ N + M++IFH + + V V+++D+++ S E
Sbjct: 804 KKTTLKINRVNYYYQVMPLGLKNI*TTYQRLMDKIFHKQI*KNVEVYVEDMIVKSSQE*F 625
Query: 678 H 678
H
Sbjct: 624 H 622
>TC12574
Length = 325
Score = 37.4 bits (85), Expect = 0.017
Identities = 15/33 (45%), Positives = 24/33 (72%)
Frame = +2
Query: 645 FMEYMNRIFHAFLDRFVVVFIDDILIYSKTEEE 677
F +N IF +F + F++VFI+DIL Y++ +EE
Sbjct: 2 FKNSVNHIFESFFEHFMIVFINDILSYTEDKEE 100
>AV779679
Length = 440
Score = 33.9 bits (76), Expect = 0.19
Identities = 15/30 (50%), Positives = 21/30 (70%)
Frame = +3
Query: 561 SMRLCIDYRQLNKVTIKNRYPLPRIDDLMD 590
+M+LC DY QL+ VTI N+ LP +D+ D
Sbjct: 345 TMQLCDDYMQLDYVTIPNKSLLPHLDEWSD 434
>TC11317 similar to UP|Q40363 (Q40363) NuM1 protein, partial (12%)
Length = 547
Score = 29.6 bits (65), Expect = 3.5
Identities = 26/78 (33%), Positives = 34/78 (43%), Gaps = 5/78 (6%)
Frame = -3
Query: 334 SFSCYYYTSATHCFIAFDCVS-----ALGLDLSDMS*EMVVETPAKGSVTTSLVCLRCPL 388
SF C+ TSA CF+A C S AL L+D+ A G+ ++LV
Sbjct: 251 SFFCFAATSAIFCFLALTCFSTSSSAALLAFLADLG------AAATGAAASTLVA----- 105
Query: 389 SMFGRDFEMDLVCLPLSG 406
DF +DL L SG
Sbjct: 104 -----DFLLDLGILTRSG 66
>TC19412 similar to UP|Q84KB0 (Q84KB0) Pol protein, partial (7%)
Length = 519
Score = 29.3 bits (64), Expect = 4.6
Identities = 17/59 (28%), Positives = 29/59 (48%)
Frame = +2
Query: 1324 EVDPEVYWSVSDIRKSWNGGLSSGFTTASFKFARRFPCVATSEVCSGSISCDPE*RCAS 1382
+ + +V+W+V +R+ W LS G + SF + F C +E+ S S * C +
Sbjct: 47 KTESKVHWTV*GVREGWLCILSFGTSARSFCSSSGFSCFYAAEIPI*SFSRYSS*GCTA 223
>TC13122 similar to UP|Q84Y01 (Q84Y01) Inositol phosphate kinase, partial
(14%)
Length = 637
Score = 29.3 bits (64), Expect = 4.6
Identities = 16/61 (26%), Positives = 29/61 (47%), Gaps = 1/61 (1%)
Frame = +3
Query: 658 DRFVVVFIDDILIYSKTEEEHAEHLKIVLQVLKEKKLYAKLSK-CEFWLKEVSFLGHVIS 716
++ + F D++ + EEE E K KEK+ ++ K C FW+ +S + +
Sbjct: 123 EKVLTEFFCDVMCQKQQEEEQGEGDKAASPAEKEKEKSVQV*KNCVFWVTRISSVDPIAG 302
Query: 717 G 717
G
Sbjct: 303 G 305
>TC17915 similar to GB|AAM10415.1|20147411|AY093799 AT3g05640/F18C1_9
{Arabidopsis thaliana;}, partial (37%)
Length = 506
Score = 28.5 bits (62), Expect = 7.9
Identities = 22/81 (27%), Positives = 37/81 (45%)
Frame = +2
Query: 596 RVFSKIDLRSGYHQIKVKDEDMQKTALRTRYGHYEYKVMPFGVTNAPRVFMEYMNRIFHA 655
RVF +D G H++ + DE+ A+ +G +Y V +G+ + P V +
Sbjct: 26 RVFC-LDDEPGVHRVCLPDEESPGLAMSRAFG--DYCVKEYGLISVPEVTQRNITS---- 184
Query: 656 FLDRFVVVFIDDILIYSKTEE 676
D+FVV+ D + EE
Sbjct: 185 -KDQFVVLATDGVWDVISNEE 244
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.360 0.161 0.597
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,051,729
Number of Sequences: 28460
Number of extensions: 349615
Number of successful extensions: 3340
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 2429
Number of HSP's successfully gapped in prelim test: 67
Number of HSP's that attempted gapping in prelim test: 849
Number of HSP's gapped (non-prelim): 2593
length of query: 1445
length of database: 4,897,600
effective HSP length: 102
effective length of query: 1343
effective length of database: 1,994,680
effective search space: 2678855240
effective search space used: 2678855240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146805.1


