

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146790.6 - phase: 0
(360 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
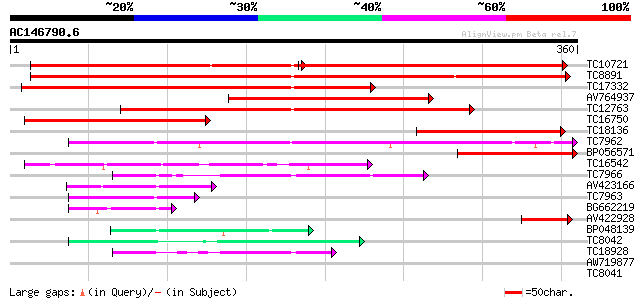
Score E
Sequences producing significant alignments: (bits) Value
TC10721 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehyd... 221 e-116
TC8891 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Ara... 343 3e-95
TC17332 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehyd... 268 8e-73
AV764937 226 6e-60
TC12763 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehyd... 206 5e-54
TC16750 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehyd... 134 2e-32
TC18136 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehyd... 88 2e-18
TC7962 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 ... 88 3e-18
BP056571 84 4e-17
TC16542 similar to UP|Q93X81 (Q93X81) Sorbitol dehydrogenase, pa... 72 1e-13
TC7966 similar to UP|Q84V25 (Q84V25) Quinone oxidoreductase, par... 62 2e-10
AV423166 59 1e-09
TC7963 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 ... 55 1e-08
BG662219 54 3e-08
AV422928 49 1e-06
BP048139 49 2e-06
TC8042 homologue to UP|P93619 (P93619) Probable NADP-dependent o... 46 8e-06
TC18928 similar to UP|Q43677 (Q43677) Auxin-induced protein, par... 43 9e-05
AW719877 37 0.004
TC8041 similar to UP|P93619 (P93619) Probable NADP-dependent oxi... 37 0.005
>TC10721 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , complete
Length = 1297
Score = 221 bits (562), Expect(2) = e-116
Identities = 106/171 (61%), Positives = 136/171 (78%)
Frame = +1
Query: 184 GKRIGVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQA 243
GK +GV GLGGLGH+A+KFGKAFG VTVISTSP+K+AEA E+LGAD F++ST+P +++A
Sbjct: 622 GKHLGVAGLGGLGHVAIKFGKAFGLKVTVISTSPNKEAEAIEKLGADSFLVSTDPAKMKA 801
Query: 244 AKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGII 303
A ++ +I+DT+SA H+L+P+L LLK+NG L VG P KPL+LP FPL+ G++ + G
Sbjct: 802 ALGTIHYIIDTISASHSLIPLLGLLKLNGKLVTVGLPGKPLELPVFPLVAGRKLVGGSHF 981
Query: 304 GGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIA 354
GGIKETQEML+ KHNI DIELIK D IN A +RL K DV+YRFVID+A
Sbjct: 982 GGIKETQEMLEFSAKHNIAADIELIKIDEINTAMERLSKADVKYRFVIDVA 1134
Score = 216 bits (549), Expect(2) = e-116
Identities = 99/175 (56%), Positives = 128/175 (72%)
Frame = +3
Query: 14 GWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEI 73
GWAA D+SG ++P+ F RRENG DV IKIL+CG+CH+D+H K+DWG T YPVVPGHEI
Sbjct: 117 GWAARDTSGVLSPFHFSRRENGDDDVAIKILFCGVCHSDLHTIKNDWGFTTYPVVPGHEI 296
Query: 74 TGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSIT 133
GV+TK G++V FK GDKVGVG + SC CE C D ENYC + + YN + +G+ T
Sbjct: 297 VGVVTKTGNNVTKFKVGDKVGVGVIVDSCKSCENCDQDLENYCPQPVYTYNSPY-NGTRT 473
Query: 134 YGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIG 188
GGYS +VV RYV+ P++LP+DA APLLCAGITV+ P+K +G+ + GK +G
Sbjct: 474 NGGYSDFVVVHQRYVLQFPDNLPLDAGAPLLCAGITVYXPMKYYGM-TEPGKTLG 635
>TC8891 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Arabidopsis
thaliana;} , complete
Length = 1636
Score = 343 bits (879), Expect = 3e-95
Identities = 173/344 (50%), Positives = 238/344 (68%), Gaps = 1/344 (0%)
Frame = +1
Query: 14 GWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEI 73
GWAA D+SG ++PY F RR G DV +KI +CG+C+ DV ++ G ++YP VPGHEI
Sbjct: 127 GWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEI 306
Query: 74 TGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEK-LQFVYNGIFWDGSI 132
G++TKVGS+V GF GD VGVG SC DCE+C E++C K F +NG+ DG+I
Sbjct: 307 AGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHDGTI 486
Query: 133 TYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGL 192
T GGYS+ +VV RY IP+S P+ +AAPLLCAGITV+SP+ H + + GK +GV+GL
Sbjct: 487 TKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKM-NQPGKSLGVIGL 663
Query: 193 GGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLDFIL 252
GGLGHMAVKFGKAFG +VTV STS SK+ EA LGAD F++S++ ++++A +SLDFI+
Sbjct: 664 GGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIV 843
Query: 253 DTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIKETQEM 312
DT S H+ P + LLK G L +VG P + +++ LI G R++ G +GG KE ++M
Sbjct: 844 DTASGPHSFDPYMALLKSFGVLALVGFPGE-IKIHPGLLIMGSRTVSGSGVGGTKEIRDM 1020
Query: 313 LDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANA 356
++ C + I +IE++ NEA R++K DV+YRFVIDI N+
Sbjct: 1021IEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENS 1152
>TC17332 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (63%)
Length = 736
Score = 268 bits (686), Expect = 8e-73
Identities = 127/225 (56%), Positives = 163/225 (72%)
Frame = +1
Query: 8 HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPV 67
H + GWAA DSSG ++P+ F RRE G DVT K+LYCGICH+D+H K++WG ++YP+
Sbjct: 64 HPKKAFGWAARDSSGVLSPFKFSRRETGEKDVTFKVLYCGICHSDLHMIKNEWGSSVYPL 243
Query: 68 VPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIF 127
VPGHEI GV+T+VGS V+ FK GD+VGVGC+ SC C+ C + ENYC K+ Y
Sbjct: 244 VPGHEIAGVVTEVGSKVQKFKVGDRVGVGCMVNSCRSCQSCADNLENYCPKMILTYAAKN 423
Query: 128 WDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRI 187
DG+ITYGGYS ++V D + + I ++LP++AAAP LCAGITV+SPLK +GL G +
Sbjct: 424 VDGTITYGGYSDLMVSDEDFGIRISDNLPLEAAAPFLCAGITVYSPLKYYGL-DKPGLHV 600
Query: 188 GVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDF 232
GV GLGGLGHMAVKF A G VT ISTSP+K+ EA E++ AD F
Sbjct: 601 GVSGLGGLGHMAVKFAIALGAKVTAISTSPNKKTEAIEKV*ADSF 735
>AV764937
Length = 392
Score = 226 bits (575), Expect = 6e-60
Identities = 109/130 (83%), Positives = 123/130 (93%)
Frame = +1
Query: 140 MLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLGGLGHMA 199
M+V DYRYVVHIPE+LPMDAAAPLLCAGITVFSPLKDH LVS+ GK+IGVVGLGGLGH+A
Sbjct: 1 MMVADYRYVVHIPENLPMDAAAPLLCAGITVFSPLKDHDLVSSPGKKIGVVGLGGLGHIA 180
Query: 200 VKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLDFILDTVSADH 259
VKFGKAFGHHVTVISTSPSK+AEAK+RLGADDFI+STN LQ+A+RSLDF+ DTVSA+H
Sbjct: 181 VKFGKAFGHHVTVISTSPSKEAEAKQRLGADDFILSTNQQHLQSARRSLDFVSDTVSAEH 360
Query: 260 ALLPILELLK 269
+LLP+LELLK
Sbjct: 361 SLLPLLELLK 390
>TC12763 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (62%)
Length = 809
Score = 206 bits (524), Expect = 5e-54
Identities = 103/225 (45%), Positives = 146/225 (64%)
Frame = +2
Query: 71 HEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDG 130
HE+ G + +VGSDV F G+ VG G L C C C++D E YC+K + YN ++ DG
Sbjct: 68 HEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDG 247
Query: 131 SITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVV 190
T GG+++ ++V+ ++VV IPE + + APLLCA +TV+SPL GL +G R G++
Sbjct: 248 KPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGL-KESGLRGGIL 424
Query: 191 GLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLDF 250
GLGG+GHM V KA GHHVTVIS+S K+ EA E LGAD +++S++ +Q A SLD+
Sbjct: 425 GLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDY 604
Query: 251 ILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGK 295
I+DTV H L P L LLK++G L ++G + PLQ ++ GK
Sbjct: 605 IIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGK 739
>TC16750 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (34%)
Length = 442
Score = 134 bits (337), Expect = 2e-32
Identities = 59/118 (50%), Positives = 78/118 (66%)
Frame = +1
Query: 10 QTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVP 69
+T GWAA D SG ++PYTF R G DV IK+ YCG+CHTDVH K+D G++ YP+VP
Sbjct: 85 RTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVP 264
Query: 70 GHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIF 127
GHE+ G + +VGSDV F G+ VG G L C C C++D E YC+K + YN ++
Sbjct: 265 GHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVY 438
>TC18136 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (28%)
Length = 601
Score = 88.2 bits (217), Expect = 2e-18
Identities = 44/95 (46%), Positives = 63/95 (66%)
Frame = +3
Query: 259 HALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIKETQEMLDVCGK 318
H L P L LLK++G L ++G PLQ ++ G+RSI G IG IKET+EM++ +
Sbjct: 9 HPLEPYLSLLKLDGKLILMGVIYTPLQFITPMVMLGRRSITGSFIGSIKETEEMVEFWKE 188
Query: 319 HNITCDIELIKADTINEAFQRLLKNDVRYRFVIDI 353
+T IE++ D I++ F+RL KNDVRYRFV+D+
Sbjct: 189 KGLTSMIEIVNMDYIHKPFERLEKNDVRYRFVVDV 293
>TC7962 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 , complete
Length = 1489
Score = 87.8 bits (216), Expect = 3e-18
Identities = 81/353 (22%), Positives = 148/353 (40%), Gaps = 30/353 (8%)
Frame = +1
Query: 38 DVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGC 97
+V +KILY +CHTDV+ + ++P + GHE G++ VG V K GD +
Sbjct: 220 EVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESVGEGVTHLKPGDH-ALPV 396
Query: 98 LAASCLDCEYCKTDQENYCEKL---------------QFVYNGIFWDGSITYGGYSQMLV 142
C +C +CK+++ N C+ L +F G + +S+ V
Sbjct: 397 FTGECGECPHCKSEESNMCDLLRINTDRGVMISDNQSRFSIKGKPIYHFVGTSTFSEYTV 576
Query: 143 VDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLGGLGHMAVKF 202
+ V I + P+D L C T F + G + + GLG +G A +
Sbjct: 577 LHAGCVAKINPAAPLDKVCILSCGICTGFGATVNVA-KPKPGSSVAIFGLGAVGLAAAEG 753
Query: 203 GKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQ------LQAAKRSLDFILDTVS 256
+ G + S + E ++ G ++F+ + D+ + +D ++
Sbjct: 754 ARVSGASRIIGVDLVSSRFEGAKKFGVNEFVNPKDHDKPVQEVIAEMTNGGVDRAVECTG 933
Query: 257 ADHALLPILELLKVN-GTLFIVGAPDKPLQLPAFPLIF-GKRSIKGGIIGGIKETQEMLD 314
+ A++ E + G +VG P++ P+ F +R++KG G K ++ +
Sbjct: 934 SIQAMISAFECVHDGWGVAVLVGVPNQDDAFQTHPVNFLNERTLKGTFYGNYKPRTDLPN 1113
Query: 315 VCGKHNITCDIELIKADT-------INEAFQRLLKNDVRYRFVIDIANASPAT 360
V + + ++EL K T IN+AF +LK + R +I + +P T
Sbjct: 1114VVEMY-MRGELELEKFITHTVSFSEINQAFDYMLKGE-SIRCIIRMEE*NPKT 1266
>BP056571
Length = 319
Score = 84.0 bits (206), Expect = 4e-17
Identities = 39/76 (51%), Positives = 55/76 (72%)
Frame = -1
Query: 285 QLPAFPLIFGKRSIKGGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKND 344
++P F L+ G+++I +IGGIKETQEM+D KH++ +IE+I D +N A +RLLK D
Sbjct: 319 EVPVFSLLGGRKTIAXSMIGGIKETQEMIDFAAKHDVKPEIEVIPMDYVNTAMERLLKAD 140
Query: 345 VRYRFVIDIANASPAT 360
V+YRFVIDI N A+
Sbjct: 139 VKYRFVIDIGNTLKAS 92
>TC16542 similar to UP|Q93X81 (Q93X81) Sorbitol dehydrogenase, partial (60%)
Length = 724
Score = 72.0 bits (175), Expect = 1e-13
Identities = 60/231 (25%), Positives = 105/231 (44%), Gaps = 10/231 (4%)
Frame = +1
Query: 10 QTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKD----DWGITMY 65
+ ++ W ++ KI P FK G DV +++ GIC +DVH+ K D+ I
Sbjct: 88 ENMAAWLVAINTLKIQP--FKLPTLGPHDVRVRMKAVGICGSDVHYLKKLRCADF-IVKE 258
Query: 66 PVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNG 125
P+V GHE G+I VG++V+ GD+V + SC C++CK N C ++
Sbjct: 259 PMVIGHECAGIIEAVGAEVKSLVPGDRVAIE-PGISCWRCDHCKLGSYNLCPDMK----- 420
Query: 126 IFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGK 185
F+ +G + +V +P+++ ++ A +C ++V G+ +
Sbjct: 421 -FFATPPVHGSLANQIVHPADLCFKLPDNVSLEEGA--MCEPLSV-------GVHACRRA 570
Query: 186 RIG------VVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGAD 230
IG ++G G +G + + +AFG VI + + LGAD
Sbjct: 571 NIGPETNVLIMGAGPIGLVTMLAARAFGAPRIVIVDVDDHRLSVAKTLGAD 723
>TC7966 similar to UP|Q84V25 (Q84V25) Quinone oxidoreductase, partial (98%)
Length = 1908
Score = 61.6 bits (148), Expect = 2e-10
Identities = 58/203 (28%), Positives = 90/203 (43%), Gaps = 2/203 (0%)
Frame = +2
Query: 66 PVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNG 125
P PG+++ GV+ KVGS+V+ FK GD+V G + + L EY K
Sbjct: 914 PTAPGYDVAGVVVKVGSEVKKFKVGDEV-YGDINENAL--EYPKV--------------- 1039
Query: 126 IFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGK 185
G ++ V+ + + H P++L AA L A T + L+ G +AGK
Sbjct: 1040 --------VGSLAEYTAVEEKLLAHKPKNLSFVEAASLPLAIETAYEGLERAGF--SAGK 1189
Query: 186 RIGVVG-LGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAA 244
I V+G GG+G ++ K V +TS + + E +LGA D I + ++
Sbjct: 1190 SILVLGGAGGVGTHVIQLAKHVFGASKVAATSSTGKLELLRKLGA-DMPIDYTKENVENL 1366
Query: 245 KRSLDFILDTV-SADHALLPILE 266
D + D V AD P+ E
Sbjct: 1367 PEKFDVVFDAVGDADTTFKPLKE 1435
>AV423166
Length = 452
Score = 58.9 bits (141), Expect = 1e-09
Identities = 29/95 (30%), Positives = 48/95 (50%)
Frame = +2
Query: 37 TDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVG 96
T+V +K+L ICHTD+ + ++P+ GHE G++ VG V K+GD + +
Sbjct: 155 TEVRVKMLCASICHTDITSIHG-FPHGIFPLALGHEGVGIVESVGDQVTNLKEGDMI-IP 328
Query: 97 CLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGS 131
C +CE C + + N C K +G+ D +
Sbjct: 329 TYIGECQECENCVSGETNLCMKYPIALSGMMPDNT 433
>TC7963 homologue to UP|Q84V97 (Q84V97) Alcohol dehydrogenase 1 , partial
(33%)
Length = 694
Score = 55.5 bits (132), Expect = 1e-08
Identities = 26/83 (31%), Positives = 44/83 (52%)
Frame = +3
Query: 38 DVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGC 97
+V +KILY +CHTDV+ + ++P + GHE G++ VG V K D +
Sbjct: 387 EVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESVGEGVTHLKPVDH-ALPV 563
Query: 98 LAASCLDCEYCKTDQENYCEKLQ 120
C + +CK+++ N C+ L+
Sbjct: 564 FTGECGELPHCKSEESNMCDLLR 632
>BG662219
Length = 366
Score = 54.3 bits (129), Expect = 3e-08
Identities = 28/71 (39%), Positives = 40/71 (55%), Gaps = 2/71 (2%)
Frame = +2
Query: 38 DVTIKILYCGICHTDVH--HAKDDWGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGV 95
+V +KILY +CHTD + KD G+ +P + GHE GV+ VG V + GD V +
Sbjct: 161 EVRVKILYTALCHTDAYTWSGKDPEGL--FPCILGHEAAGVVESVGEGVTEVQPGDHV-I 331
Query: 96 GCLAASCLDCE 106
C A C +C+
Sbjct: 332 PCYQAECRECK 364
>AV422928
Length = 399
Score = 49.3 bits (116), Expect = 1e-06
Identities = 24/32 (75%), Positives = 26/32 (81%)
Frame = -3
Query: 326 ELIKADTINEAFQRLLKNDVRYRFVIDIANAS 357
ELI D INEA +RL KNDVRYRFVIDIA +S
Sbjct: 397 ELITPDRINEAMERLAKNDVRYRFVIDIAGSS 302
>BP048139
Length = 567
Score = 48.5 bits (114), Expect = 2e-06
Identities = 40/144 (27%), Positives = 56/144 (38%), Gaps = 15/144 (10%)
Frame = -2
Query: 65 YPVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQF-VY 123
+P + GHE GV+ VG DV GD V + C +C CK+ + N C F V
Sbjct: 431 FPRILGHEAVGVVESVGKDVTEVTKGDVV-IPIYLPDCGECTDCKSTKSNRCTNFPFKVS 255
Query: 124 NGIFWDGSITY--------------GGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGIT 169
+ DG+ + +S+ VVD V I +P D A L C GI+
Sbjct: 254 PSMPRDGTTRFTDLNGEIIHHFMFISSFSEYTVVDVANVTKIDPEIPPDRACLLSC-GIS 78
Query: 170 VFSPLKDHGLVSTAGKRIGVVGLG 193
G + + GLG
Sbjct: 77 TGVGAAWRTASVEPGSTVAIFGLG 6
>TC8042 homologue to UP|P93619 (P93619) Probable NADP-dependent
oxidoreductase TED2 , complete
Length = 1244
Score = 46.2 bits (108), Expect = 8e-06
Identities = 46/188 (24%), Positives = 70/188 (36%)
Frame = +2
Query: 38 DVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGC 97
+V +K G+ DV+ K + + P PG E GV+T VG G + GD VG
Sbjct: 203 EVRVKNKAIGVNFIDVYFRKGVYKASTSPFTPGMEAVGVVTAVGGGPTGVQVGDLVG--- 373
Query: 98 LAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSITYGGYSQMLVVDYRYVVHIPESLPM 157
Y G G Y + ++ VV IP S+
Sbjct: 374 -------------------------YAG------QPIGSYVEEQILHAGKVVPIPSSIDP 460
Query: 158 DAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLGGLGHMAVKFGKAFGHHVTVISTSP 217
AA ++ G+T L+ V + GG+G + ++ A G V ++
Sbjct: 461 SVAASVMLKGMTAQFLLRRCFKVERGHTILVHAAAGGVGSLLCQWANALGATVIGTVSNE 640
Query: 218 SKQAEAKE 225
K A+AKE
Sbjct: 641 EKAAQAKE 664
>TC18928 similar to UP|Q43677 (Q43677) Auxin-induced protein, partial (53%)
Length = 612
Score = 42.7 bits (99), Expect = 9e-05
Identities = 38/143 (26%), Positives = 64/143 (44%), Gaps = 1/143 (0%)
Frame = +1
Query: 66 PVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNG 125
P PG+++ GV+ KVGS+V+ FK GD+V Y +++ VY
Sbjct: 268 PTAPGYDVAGVVVKVGSEVKKFKVGDEV-------------YGDINEQ------AMVYPK 390
Query: 126 IFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGK 185
+ G ++ + + + H P++L AA L T ++ L+ G +AGK
Sbjct: 391 V-------VGSLAEYTAAEEKLLSHKPQNLSFAEAASLPLTIETAYNGLELAGF--SAGK 543
Query: 186 RIGVV-GLGGLGHMAVKFGKAFG 207
I V+ G GG+G + +G
Sbjct: 544 SILVLGGAGGVGTSLFSWQACYG 612
>AW719877
Length = 520
Score = 37.4 bits (85), Expect = 0.004
Identities = 32/119 (26%), Positives = 51/119 (41%), Gaps = 6/119 (5%)
Frame = +2
Query: 126 IFWDGSIT-YGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDH-----GL 179
+F+ G++ G ++ +VD R V P SL AA L IT F L D +
Sbjct: 116 VFYSGALAPQGTNAEFHLVDERLVGRKPSSLDYAQAAALPLTAITAFEALFDRLDVKKPV 295
Query: 180 VSTAGKRIGVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNP 238
V A + + G GG+G +AV+ + + + S + + LGA I + P
Sbjct: 296 VGAANAILIIGGAGGVGSIAVQLARQLTDLTVIATASRPETRDWVLGLGAHHVIDHSKP 472
>TC8041 similar to UP|P93619 (P93619) Probable NADP-dependent
oxidoreductase TED2 , partial (31%)
Length = 637
Score = 37.0 bits (84), Expect = 0.005
Identities = 20/57 (35%), Positives = 28/57 (49%)
Frame = +1
Query: 38 DVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVG 94
+V +K G+ DV+ K + + P PG E GV+T VG G + GD VG
Sbjct: 427 EVRVKNKAIGVNFIDVYFRKGVYKASSTPFTPGMEAVGVVTAVGGGPTGVQVGDLVG 597
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,280,792
Number of Sequences: 28460
Number of extensions: 87307
Number of successful extensions: 396
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 379
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 381
length of query: 360
length of database: 4,897,600
effective HSP length: 91
effective length of query: 269
effective length of database: 2,307,740
effective search space: 620782060
effective search space used: 620782060
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146790.6


