

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146784.6 - phase: 0
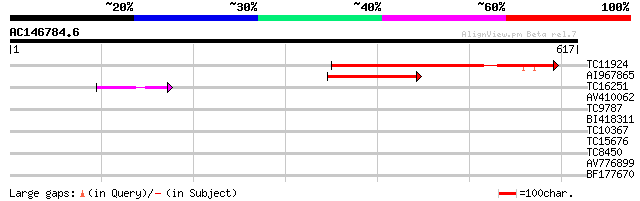
(617 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC11924 similar to UP|P93484 (P93484) BP-80 vacuolar sorting rec... 281 3e-76
AI967865 162 1e-40
TC16251 similar to UP|Q945B9 (Q945B9) Growth-on protein GRO11, p... 50 8e-07
AV410062 30 1.5
TC9787 28 4.3
BI418311 28 4.3
TC10367 28 4.3
TC15676 similar to GB|AAG24281.1|10880503|AF195894 arabinogalact... 27 7.4
TC8450 similar to UP|PDA1_VIGUN (O04865) Phospholipase D alpha 1... 27 7.4
AV776899 27 9.7
BF177670 27 9.7
>TC11924 similar to UP|P93484 (P93484) BP-80 vacuolar sorting receptor,
partial (41%)
Length = 783
Score = 281 bits (718), Expect = 3e-76
Identities = 140/273 (51%), Positives = 178/273 (64%), Gaps = 26/273 (9%)
Frame = +3
Query: 351 NPVLKAEQEAQIGKESRGDVTILPTLVINNRQYRGKLSRPAVLKAMCAGFQETTEPSICL 410
+PVLK EQ+AQ+GK SRGDVTILPTLV+NNRQYRGKL + AV+KA+C+GF+ETTEP++CL
Sbjct: 3 DPVLKEEQDAQVGKGSRGDVTILPTLVVNNRQYRGKLEKGAVMKAICSGFEETTEPAVCL 182
Query: 411 TPDMETNECLENNGGCWKEKSSNITACRDTFRGRVCVCPVVNNIKFVGDGYTHCEASGTL 470
+ D+ETNECLENNGGCWK+K++NITAC+DTFRGRVC CP+V+ ++F GDGYT CEASG
Sbjct: 183 SSDVETNECLENNGGCWKDKAANITACKDTFRGRVCECPLVDGVQFKGDGYTTCEASGPG 362
Query: 471 SCEFNNGGCWKASHGGRLYSACHDDYRKGCECPSGFRGDGVRSCEDCFNIFRILTMLQIL 530
C+ NGGCW + G +SAC DD C+CP+GF+GDGV++C+D +
Sbjct: 363 RCKIKNGGCWHEARNGHAFSACVDDGEVKCQCPAGFKGDGVKNCQD-------------V 503
Query: 531 MNAKKSRLASVHNANAKIPLGVMSASA-----------IAVCSTHEKMTR---------- 569
K+ + + K G S + T + R
Sbjct: 504 DECKEKKACQCPECSCKNTWGSYDCSCSGDLLYIRDHDTCISKTASQGGRSAWAAFWVII 683
Query: 570 ---VLS--GGYAFYKYRIQRYMDTEIRAIMAQY 597
VLS GGY YKYRI+ YMD+EIRAIMAQY
Sbjct: 684 AGLVLSAGGGYLIYKYRIRSYMDSEIRAIMAQY 782
>AI967865
Length = 326
Score = 162 bits (410), Expect = 1e-40
Identities = 73/102 (71%), Positives = 89/102 (86%)
Frame = +2
Query: 347 ADVENPVLKAEQEAQIGKESRGDVTILPTLVINNRQYRGKLSRPAVLKAMCAGFQETTEP 406
ADVEN VLK EQ+ Q+G+ SRGDVTILPTLVINN QY GKL R VLKA+CAGF+E+TEP
Sbjct: 20 ADVENEVLKNEQQVQVGRGSRGDVTILPTLVINNVQYTGKLERTPVLKAVCAGFKESTEP 199
Query: 407 SICLTPDMETNECLENNGGCWKEKSSNITACRDTFRGRVCVC 448
+CL+ D+ETNECLE NGGCW++K+++I+AC+DTFRGRVC C
Sbjct: 200 PVCLSGDIETNECLERNGGCWQDKNASISACKDTFRGRVCEC 325
>TC16251 similar to UP|Q945B9 (Q945B9) Growth-on protein GRO11, partial (9%)
Length = 905
Score = 50.4 bits (119), Expect = 8e-07
Identities = 31/83 (37%), Positives = 44/83 (52%)
Frame = +3
Query: 95 LVDRGDCYFTLKAWNAQNGGAAAILVADDREETLITMDTPEEGNVVNDDYIEKINIPSAL 154
L RG C FT KA AQ+GGA+A+L+ +D E+ + G I+IP L
Sbjct: 312 LCTRGGCDFTTKAAFAQSGGASAMLLINDEEDLFEMACSNSTGG--------NISIPVVL 467
Query: 155 ISKSLGDRIKKALSDGEMVHINL 177
I+KS G+ K+L+ G V + L
Sbjct: 468 ITKSAGEAFNKSLASGRKVEVLL 536
>AV410062
Length = 435
Score = 29.6 bits (65), Expect = 1.5
Identities = 13/27 (48%), Positives = 17/27 (62%)
Frame = -3
Query: 477 GGCWKASHGGRLYSACHDDYRKGCECP 503
GGCW+ S+GG +SA R+G E P
Sbjct: 397 GGCWEKSNGGNTFSAA----RRGTENP 329
>TC9787
Length = 716
Score = 28.1 bits (61), Expect = 4.3
Identities = 23/96 (23%), Positives = 38/96 (38%)
Frame = +1
Query: 466 ASGTLSCEFNNGGCWKASHGGRLYSACHDDYRKGCECPSGFRGDGVRSCEDCFNIFRILT 525
A+G L G C + G + C D ++GC R FN+ L
Sbjct: 253 AAGALIATGQGGNCPPGTGGRDRSTNCFDQVKRGCHHDPYLRSGC**MLAVIFNMSSSLL 432
Query: 526 MLQILMNAKKSRLASVHNANAKIPLGVMSASAIAVC 561
+L +L+ ++ + AS +P V S+ I+ C
Sbjct: 433 LLHLLLFSRIGKFASF----PILPRFVCSSLCISTC 528
>BI418311
Length = 540
Score = 28.1 bits (61), Expect = 4.3
Identities = 13/34 (38%), Positives = 20/34 (58%)
Frame = -2
Query: 528 QILMNAKKSRLASVHNANAKIPLGVMSASAIAVC 561
+ILM A+ L S+ N + + ++S SA AVC
Sbjct: 179 RILMTAEVESLKSISAVNLSLSMSMLSISAAAVC 78
>TC10367
Length = 527
Score = 28.1 bits (61), Expect = 4.3
Identities = 16/39 (41%), Positives = 20/39 (51%), Gaps = 3/39 (7%)
Frame = +3
Query: 486 GRLYSACHDDYRKGCE---CPSGFRGDGVRSCEDCFNIF 521
GR Y C +++ C C SG G GVR+ E CF F
Sbjct: 117 GRHYI*CICGWKRRCSWNCCGSGMVGCGVRN*EPCFEDF 233
>TC15676 similar to GB|AAG24281.1|10880503|AF195894 arabinogalactan protein
{Arabidopsis thaliana;} , partial (49%)
Length = 877
Score = 27.3 bits (59), Expect = 7.4
Identities = 12/37 (32%), Positives = 17/37 (45%), Gaps = 3/37 (8%)
Frame = +2
Query: 469 TLSCEFNNGGCWKASHGG---RLYSACHDDYRKGCEC 502
T C F NGG W+ S G + + C+ + C C
Sbjct: 113 TSGCRFGNGGEWRLSR*GTSSKSHIGCYYSFCSSCFC 223
>TC8450 similar to UP|PDA1_VIGUN (O04865) Phospholipase D alpha 1 (PLD alpha
1) (Choline phosphatase 1)
(Phosphatidylcholine-hydrolyzing phospholipase D 1) ,
partial (73%)
Length = 2062
Score = 27.3 bits (59), Expect = 7.4
Identities = 17/68 (25%), Positives = 33/68 (48%), Gaps = 3/68 (4%)
Frame = +2
Query: 216 FKGAAQLLEKKGFTQFTPHYITWYCPKEFLLSRRCKSQCINHGRY---CAPDPEQDFNKG 272
+K Q L KG + +Y+T++C L +R K Q G Y P+P+ D+++
Sbjct: 1148 YKDIIQALRAKGIEEDPRNYLTFFC----LGNREVKKQ----GEYEPSEQPEPDSDYSRA 1303
Query: 273 YDGKDVVV 280
+ + ++
Sbjct: 1304 QEARRFMI 1327
>AV776899
Length = 396
Score = 26.9 bits (58), Expect = 9.7
Identities = 8/18 (44%), Positives = 12/18 (66%)
Frame = +1
Query: 446 CVCPVVNNIKFVGDGYTH 463
C+C + ++ VGD YTH
Sbjct: 7 CLCTITATVRLVGDRYTH 60
>BF177670
Length = 341
Score = 26.9 bits (58), Expect = 9.7
Identities = 9/28 (32%), Positives = 18/28 (64%)
Frame = -3
Query: 235 YITWYCPKEFLLSRRCKSQCINHGRYCA 262
++T+ P +F L++ CK + GR+C+
Sbjct: 195 HVTYLHPLQFWLAKLCKHSRLRSGRHCS 112
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.138 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,386,943
Number of Sequences: 28460
Number of extensions: 193413
Number of successful extensions: 941
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 930
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 939
length of query: 617
length of database: 4,897,600
effective HSP length: 96
effective length of query: 521
effective length of database: 2,165,440
effective search space: 1128194240
effective search space used: 1128194240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146784.6


