

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146777.12 + phase: 0
(238 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
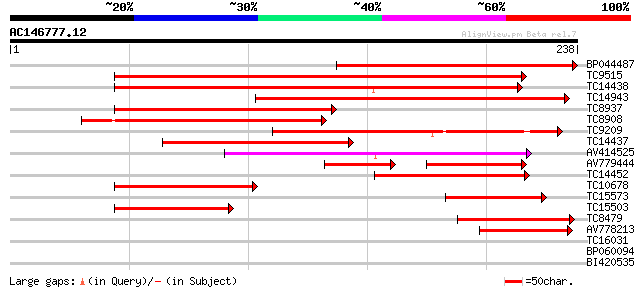
Score E
Sequences producing significant alignments: (bits) Value
BP044487 184 9e-48
TC9515 similar to UP|O49081 (O49081) Clp-like energy-dependent p... 147 2e-36
TC14438 UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease proteo... 145 6e-36
TC14943 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protea... 125 8e-30
TC8937 similar to UP|Q8W4Z1 (Q8W4Z1) ClpP (ATP-dependent Clp pr... 91 2e-19
TC8908 similar to UP|Q8LAN0 (Q8LAN0) ATP-dependent Clp protease ... 87 3e-18
TC9209 homologue to UP|Q7X9M5 (Q7X9M5) ATP-dependent Clp proteas... 85 1e-17
TC14437 UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease proteo... 76 5e-15
AV414525 71 1e-13
AV779444 43 8e-10
TC14452 similar to UP|Q94B60 (Q94B60) ATP-dependent Clp protease... 54 2e-08
TC10678 similar to PIR|T52454|T52454 ATP-dependent Clp proteinas... 52 1e-07
TC15573 similar to UP|O82421 (O82421) ATP-dependent Clp protease... 42 1e-04
TC15503 similar to UP|O04018 (O04018) F7G19.1 protein, partial (... 41 2e-04
TC8479 similar to UP|O04018 (O04018) F7G19.1 protein, partial (9%) 39 6e-04
AV778213 39 8e-04
TC16031 similar to UP|O23548 (O23548) CLP proteinase homolog (CL... 29 0.82
BP060094 28 1.8
BI420535 25 9.1
>BP044487
Length = 533
Score = 184 bits (468), Expect = 9e-48
Identities = 87/101 (86%), Positives = 96/101 (94%)
Frame = -1
Query: 138 RRALPNATIMIHQPSGGYSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDR 197
RR+LPNA+IMIHQPSGGYSGQAKDIAIHTKQIVR+WD LN+LY KHTGQ +++IQKNMDR
Sbjct: 533 RRSLPNASIMIHQPSGGYSGQAKDIAIHTKQIVRVWDLLNDLYAKHTGQSIELIQKNMDR 354
Query: 198 DYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVADEGKDKGSN 238
DYFMT EEAKEFG+IDEVIDQRPMTLVSDAV +EGKDKGSN
Sbjct: 353 DYFMTPEEAKEFGLIDEVIDQRPMTLVSDAVGNEGKDKGSN 231
>TC9515 similar to UP|O49081 (O49081) Clp-like energy-dependent protease
(ATP-dependent Clp protease proteolytic subunit)
(Endopeptidase Clp) , partial (74%)
Length = 1158
Score = 147 bits (371), Expect = 2e-36
Identities = 70/173 (40%), Positives = 110/173 (63%)
Frame = +1
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
D+ S L + RI+ I P++ A V++QL+ L + +P+ I MY+N PGG+ + LAIY
Sbjct: 310 DLSSVLFRNRIIFIGQPVNAQVAQRVISQLVTLATIDPNADILMYINCPGGSTYSVLAIY 489
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAI 164
D M +I+ + T+C G AAS G+LLL G KG R ++PN+ IMIHQP G G +D+
Sbjct: 490 DCMSWIKPKVGTVCFGVAASQGALLLAGGEKGMRYSMPNSRIMIHQPQSGCGGHVEDVRR 669
Query: 165 HTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVID 217
+ V+ ++++Y TGQP++ +Q+ +RD F++ EA EFG+ID V++
Sbjct: 670 QVNEAVQSRHKIDKMYSAFTGQPIEKVQQYTERDRFLSVSEALEFGLIDGVLE 828
>TC14438 UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease proteolytic
subunit (Endopeptidase Clp) , complete
Length = 620
Score = 145 bits (366), Expect = 6e-36
Identities = 64/172 (37%), Positives = 118/172 (68%), Gaps = 1/172 (0%)
Frame = +3
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
DI++RL +ER++ + ++ + ++ ++ +++L E+ K + +++NSPGG V G+AIY
Sbjct: 93 DIYNRLYRERLLFLGQEVNSEISNQLIGLMVYLSIEDDKKDLYLFINSPGGWVIPGIAIY 272
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQP-SGGYSGQAKDIA 163
DTMQ+++ + T+C+G AASMGS +L G +R A P+A +MIHQP S Y Q +
Sbjct: 273 DTMQFVQPDVQTVCMGLAASMGSFVLAGGKITKRLAFPHARVMIHQPASSFYEAQTGEFI 452
Query: 164 IHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEV 215
+ ++++++ + + +YV+ TG+PL ++ ++M+RD FM+A EA+ +GI+D V
Sbjct: 453 LEAEELLKLRETITRVYVQRTGKPLWLVSEDMERDVFMSAAEAQAYGIVDLV 608
>TC14943 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (ATP-dependent Clp protease proteolytic subunit
ClpP5) (Endopeptidase Clp) , partial (42%)
Length = 639
Score = 125 bits (313), Expect = 8e-30
Identities = 64/132 (48%), Positives = 88/132 (66%)
Frame = +1
Query: 104 YDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIA 163
+DTM++IR ++T+C+G AASMG+ LL AG KG+R +LPN+ IMIHQP GG G DI
Sbjct: 1 FDTMRHIRPDVSTVCVGLAASMGAFLLSAGTKGKRYSLPNSRIMIHQPLGGAQGGQTDID 180
Query: 164 IHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTL 223
I +++ LN HTGQ L+ I ++ DRD+FM+A+EAKE+G+ID VI L
Sbjct: 181 IQANEMLHHKANLNGYLSYHTGQSLEKINQDTDRDFFMSAKEAKEYGLIDGVIMNPLKAL 360
Query: 224 VSDAVADEGKDK 235
A+EGKD+
Sbjct: 361 QPLPAAEEGKDR 396
>TC8937 similar to UP|Q8W4Z1 (Q8W4Z1) ClpP (ATP-dependent Clp protease
proteolytic subunit) (Endopeptidase Clp) (Fragment) ,
partial (40%)
Length = 568
Score = 90.5 bits (223), Expect = 2e-19
Identities = 42/93 (45%), Positives = 66/93 (70%)
Frame = +3
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
D + LL++RIV + + D TA +++QLLFL++E+ K I +++NSPGG+VTAG+ IY
Sbjct: 288 DTTNMLLRQRIVFLGSQVDDMTADFIISQLLFLDAEDSKKDIKLFINSPGGSVTAGMGIY 467
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQ 137
D M+ ++ + T LG AASMG+ +L +G KG+
Sbjct: 468 DAMKLCKADVPTGRLGLAASMGAFILASGTKGK 566
>TC8908 similar to UP|Q8LAN0 (Q8LAN0) ATP-dependent Clp protease
proteolytic subunit (ClpP4), partial (35%)
Length = 598
Score = 87.0 bits (214), Expect = 3e-18
Identities = 45/103 (43%), Positives = 67/103 (64%)
Frame = +1
Query: 31 PMVIETSSRGERAYDIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYL 90
P T+ RG D LL+ERIV + I D A +++QLL L++++PSK I +++
Sbjct: 289 PQTPATAIRGAET-DAMGLLLRERIVFLGSGIDDFVADAIMSQLLLLDAQDPSKDIKLFI 465
Query: 91 NSPGGAVTAGLAIYDTMQYIRSPINTICLGQAASMGSLLLCAG 133
NSPGG+++A +AIYD Q +R+ +NTI LG AAS S++L G
Sbjct: 466 NSPGGSLSATMAIYDADQLVRADVNTIALGIAASTDSIILGGG 594
>TC9209 homologue to UP|Q7X9M5 (Q7X9M5) ATP-dependent Clp protease
proteolytic subunit (Fragment), complete
Length = 595
Score = 84.7 bits (208), Expect = 1e-17
Identities = 48/123 (39%), Positives = 75/123 (60%), Gaps = 1/123 (0%)
Frame = +2
Query: 111 RSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAIHTKQIV 170
+SP+ T C+G A ++ + LL AG KG R A+P + I + P+G GQA DI +++
Sbjct: 2 QSPVATHCVGYAYNLAAFLLAAGLKGNRYAMPLSRIALQSPAGAARGQADDIQNEANELL 181
Query: 171 RMWDAL-NELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVA 229
R+ D L NEL K TGQP++ I +++ R A+EA E+G+ID ++ RP + +DA
Sbjct: 182 RIRDYLFNELATK-TGQPVEKITQDLSRMKRFNAQEALEYGLIDRIV--RPPRIKADAPV 352
Query: 230 DEG 232
+G
Sbjct: 353 KDG 361
>TC14437 UP|CLPP_LOTJA (Q9BBQ9) ATP-dependent Clp protease proteolytic
subunit (Endopeptidase Clp) , partial (58%)
Length = 1013
Score = 76.3 bits (186), Expect = 5e-15
Identities = 33/80 (41%), Positives = 55/80 (68%)
Frame = +2
Query: 65 DTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQYIRSPINTICLGQAAS 124
+ ++ ++ +++L E+ K + +++NSPGG V G+AIYDTMQ+++ + T+C+G AAS
Sbjct: 2 EISNQLIGLMVYLSIEDDKKDLYLFINSPGGWVIPGIAIYDTMQFVQPDVQTVCMGLAAS 181
Query: 125 MGSLLLCAGAKGQRRALPNA 144
MGS +L G QR A P+A
Sbjct: 182 MGSFVLAGGKITQRLAFPHA 241
>AV414525
Length = 427
Score = 71.2 bits (173), Expect = 1e-13
Identities = 41/130 (31%), Positives = 68/130 (51%), Gaps = 1/130 (0%)
Frame = +3
Query: 91 NSPGGAVTAGLAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQ 150
N G+ T +I D M Y+++ + T+ G A ++LL GAKG R PN++ ++
Sbjct: 21 NETVGSETEAYSIADMMSYVKADVYTVNCGMAYGQAAMLLSLGAKGYRALQPNSSTKLYL 200
Query: 151 PS-GGYSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEF 209
P SG A D+ I K++ D EL K G+ + + K++ R + +EA E+
Sbjct: 201 PKVNRSSGAAIDMWIKAKELEANSDYYIELLAKGIGKSKEEVAKDVQRPRYFQPKEAIEY 380
Query: 210 GIIDEVIDQR 219
GI D++ID +
Sbjct: 381 GIADKIIDSQ 410
>AV779444
Length = 539
Score = 42.7 bits (99), Expect(2) = 8e-10
Identities = 18/42 (42%), Positives = 28/42 (65%)
Frame = -2
Query: 176 LNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVID 217
L Y TGQP++ +Q+ +RD F++ EA EFG+ID V++
Sbjct: 409 LTRCYSAFTGQPIEKVQQYTERDRFLSVSEALEFGLIDGVLE 284
Score = 35.8 bits (81), Expect(2) = 8e-10
Identities = 14/30 (46%), Positives = 20/30 (66%)
Frame = -1
Query: 133 GAKGQRRALPNATIMIHQPSGGYSGQAKDI 162
G KG R ++PN+ IMIH+P G G +D+
Sbjct: 539 GEKGMRYSMPNSRIMIHRPPTGCGGHVEDV 450
>TC14452 similar to UP|Q94B60 (Q94B60) ATP-dependent Clp protease-like
protein (ATP-dependent Clp protease proteolytic
subunit) (Endopeptidase Clp) (At5g45390) , partial (36%)
Length = 621
Score = 53.9 bits (128), Expect = 2e-08
Identities = 26/65 (40%), Positives = 42/65 (64%)
Frame = +2
Query: 154 GYSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIID 213
G SGQA D+ I K+++ + + + TG+ + +QK++DRD +M+ EA E+GIID
Sbjct: 2 GASGQAIDVEIQAKEVMHNKNNVTRIISGFTGRTFEQVQKDIDRDRYMSPIEAVEYGIID 181
Query: 214 EVIDQ 218
VID+
Sbjct: 182 GVIDK 196
>TC10678 similar to PIR|T52454|T52454 ATP-dependent Clp proteinase
catalytic chain P2 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(37%)
Length = 516
Score = 51.6 bits (122), Expect = 1e-07
Identities = 22/60 (36%), Positives = 42/60 (69%)
Frame = +3
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
DI++ L +ER++ I I ++ ++ ++A +L+L+S + +K + MY+N PGG +T +AIY
Sbjct: 336 DIWNALYRERVIFIGQEIDEEFSNQILATMLYLDSVDNAKKLYMYINGPGGDLTPTMAIY 515
>TC15573 similar to UP|O82421 (O82421) ATP-dependent Clp protease
proteolytic subunit (Endopeptidase Clp) (Fragment) ,
partial (25%)
Length = 567
Score = 41.6 bits (96), Expect = 1e-04
Identities = 19/42 (45%), Positives = 29/42 (68%)
Frame = +1
Query: 184 TGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVS 225
TG+P + I+ + DRD FM EAKE+G++D VID+ L++
Sbjct: 10 TGKPEEQIELDTDRDNFMNPWEAKEYGLVDGVIDEGKPGLIA 135
>TC15503 similar to UP|O04018 (O04018) F7G19.1 protein, partial (45%)
Length = 590
Score = 40.8 bits (94), Expect = 2e-04
Identities = 20/50 (40%), Positives = 32/50 (64%)
Frame = +1
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPG 94
D+ S LL RIV I P+ ++VA+L++L+ +P +PI +Y+NS G
Sbjct: 409 DLPSLLLHGRIVYIGMPLVPAVTELIVAELMYLQWMDPKEPIYIYINSTG 558
>TC8479 similar to UP|O04018 (O04018) F7G19.1 protein, partial (9%)
Length = 544
Score = 39.3 bits (90), Expect = 6e-04
Identities = 17/49 (34%), Positives = 30/49 (60%)
Frame = +1
Query: 189 DVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVADEGKDKGS 237
+ + M R Y+M + AKEFG+ID+++ + +++D A E DKG+
Sbjct: 7 ETVANVMKRPYYMDSTRAKEFGVIDQILWRGQEKVMADVAAPEEWDKGA 153
>AV778213
Length = 600
Score = 38.9 bits (89), Expect = 8e-04
Identities = 20/39 (51%), Positives = 26/39 (66%)
Frame = -3
Query: 198 DYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVADEGKDKG 236
D+FM+A+EAKE+G+ID VI L A+EGKD G
Sbjct: 598 DFFMSAKEAKEYGLIDGVIMNPL*ALQPLPAAEEGKDTG 482
>TC16031 similar to UP|O23548 (O23548) CLP proteinase homolog (CLP
proteinase like protein), partial (10%)
Length = 601
Score = 28.9 bits (63), Expect = 0.82
Identities = 15/49 (30%), Positives = 26/49 (52%)
Frame = +2
Query: 182 KHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVAD 230
KH + + I+ ++ R + + EA E+GIID+V+ T V+D
Sbjct: 5 KHIEKSPEQIEADIQRPKYFSPSEAVEYGIIDKVLYNERGTADHGVVSD 151
>BP060094
Length = 369
Score = 27.7 bits (60), Expect = 1.8
Identities = 15/42 (35%), Positives = 24/42 (56%), Gaps = 3/42 (7%)
Frame = +3
Query: 62 ISDDTAHVVVAQLLFLESENPSKPINMYLN---SPGGAVTAG 100
+SD +H ++LFL ENP++ +++ N SP G T G
Sbjct: 33 MSDQESHSHHTKVLFLTVENPNQQLHISFNKYMSPFGGCTPG 158
>BI420535
Length = 232
Score = 25.4 bits (54), Expect = 9.1
Identities = 12/29 (41%), Positives = 16/29 (54%)
Frame = +2
Query: 61 PISDDTAHVVVAQLLFLESENPSKPINMY 89
P S + A LLFL SEN S+ +M+
Sbjct: 14 PSSSSPVKTIAASLLFLSSENGSEVPSMF 100
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.135 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,376,198
Number of Sequences: 28460
Number of extensions: 39761
Number of successful extensions: 194
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 192
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 192
length of query: 238
length of database: 4,897,600
effective HSP length: 87
effective length of query: 151
effective length of database: 2,421,580
effective search space: 365658580
effective search space used: 365658580
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC146777.12


