

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146776.3 - phase: 0
(286 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
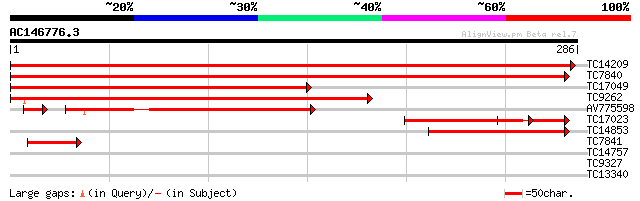
Score E
Sequences producing significant alignments: (bits) Value
TC14209 similar to UP|AAQ72788 (AAQ72788) Hypersensitive-induced... 476 e-135
TC7840 similar to UP|AAQ72788 (AAQ72788) Hypersensitive-induced ... 462 e-131
TC17049 similar to UP|Q9FM19 (Q9FM19) Hypersensitive-induced res... 269 5e-73
TC9262 similar to UP|Q9FHM7 (Q9FHM7) Gb|AAF03497.1 (At5g51570), ... 198 1e-51
AV775598 125 3e-30
TC17023 homologue to UP|Q9FM19 (Q9FM19) Hypersensitive-induced r... 101 2e-22
TC14853 similar to UP|PZ12_LUPPO (P16148) PPLZ12 protein, partia... 95 1e-20
TC7841 homologue to UP|1433_OENHO (P29307) 14-3-3-like protein, ... 46 8e-06
TC14757 similar to UP|O04361 (O04361) Prohibitin, complete 38 0.002
TC9327 similar to UP|Q38921 (Q38921) GTP-binding protein ATGB1 (... 26 6.8
TC13340 similar to UP|Q94A50 (Q94A50) AT3g55070/T15C9_70, partia... 26 8.9
>TC14209 similar to UP|AAQ72788 (AAQ72788) Hypersensitive-induced response
protein, complete
Length = 1298
Score = 476 bits (1226), Expect = e-135
Identities = 238/285 (83%), Positives = 264/285 (92%)
Frame = +1
Query: 1 MGNLVCCVQVDQSQVAMKEGFGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCET 60
MG + CVQVDQS VA+KE FGKF ++L+PGCHC+PW LG +IAG LSLRVQQLD++CET
Sbjct: 64 MGQALGCVQVDQSNVAIKEHFGKFSEILEPGCHCLPWCLGYQIAGGLSLRVQQLDVRCET 243
Query: 61 KTKDNVFVNVVASIQYRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQ 120
KTKDNVFV VVAS+QYRA+ADKA+DAFY+L+NTR QIQ+YVFDVIRASVPKL LD FEQ
Sbjct: 244 KTKDNVFVTVVASVQYRAVADKASDAFYRLTNTREQIQSYVFDVIRASVPKLELDSVFEQ 423
Query: 121 KNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEA 180
KN+IAK+VEEELEKAMS YGYEIVQTLI DIEPDV+VKRAMNEINAAARMRLAA EKAEA
Sbjct: 424 KNDIAKSVEEELEKAMSTYGYEIVQTLIVDIEPDVNVKRAMNEINAAARMRLAANEKAEA 603
Query: 181 EKILQIKRAEGEAESKYLSGLGIARQRQAIVDGLRDSVIGFSVNVPGTTAKDVMDMVLVT 240
EKILQIK+AEGEAESKYLSGLGIARQRQAIVDGLRDSV+ FS NVPGT+AKDVMDMVLVT
Sbjct: 604 EKILQIKKAEGEAESKYLSGLGIARQRQAIVDGLRDSVLAFSENVPGTSAKDVMDMVLVT 783
Query: 241 QYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQIRDGLLQGSLSH 285
QYFDTMKEIGA+SKSS+VFIPHGPGAVRD+A QIRDGLLQG+ +H
Sbjct: 784 QYFDTMKEIGASSKSSSVFIPHGPGAVRDIAVQIRDGLLQGNAAH 918
>TC7840 similar to UP|AAQ72788 (AAQ72788) Hypersensitive-induced response
protein, complete
Length = 1585
Score = 462 bits (1189), Expect = e-131
Identities = 230/282 (81%), Positives = 260/282 (91%)
Frame = +1
Query: 1 MGNLVCCVQVDQSQVAMKEGFGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCET 60
MG + CVQVDQS VA+KE FGK++ VLQPGCHC+PW +G +I+G LSLRV+QLD++CET
Sbjct: 244 MGQTLGCVQVDQSSVAIKEVFGKYDDVLQPGCHCVPWCIGSQISGSLSLRVKQLDVRCET 423
Query: 61 KTKDNVFVNVVASIQYRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQ 120
KTKDNVFV VVASIQYRALADKA DA+YKLS+T+ QIQAYVFDVIRASVPK+ LD FEQ
Sbjct: 424 KTKDNVFVTVVASIQYRALADKAVDAYYKLSDTKAQIQAYVFDVIRASVPKMELDSAFEQ 603
Query: 121 KNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEA 180
KNEIAKAVEEELEKAMSAYGYEIVQTLI DIEPD VK+AMNEINAAAR+R+A KEKAEA
Sbjct: 604 KNEIAKAVEEELEKAMSAYGYEIVQTLIVDIEPDERVKKAMNEINAAARLRVATKEKAEA 783
Query: 181 EKILQIKRAEGEAESKYLSGLGIARQRQAIVDGLRDSVIGFSVNVPGTTAKDVMDMVLVT 240
EKILQIKRAEG+AESKYL+GLGIARQRQAIVDGLRDSV+ FS NVPGT++KD+MDMVLVT
Sbjct: 784 EKILQIKRAEGDAESKYLAGLGIARQRQAIVDGLRDSVLAFSENVPGTSSKDIMDMVLVT 963
Query: 241 QYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQIRDGLLQGS 282
QYFDTMKEIGA++KS+AVFIPHGPGAV+D+ SQIRDGLLQG+
Sbjct: 964 QYFDTMKEIGASAKSNAVFIPHGPGAVKDITSQIRDGLLQGN 1089
>TC17049 similar to UP|Q9FM19 (Q9FM19) Hypersensitive-induced response
protein, partial (53%)
Length = 602
Score = 269 bits (687), Expect = 5e-73
Identities = 129/152 (84%), Positives = 143/152 (93%)
Frame = +3
Query: 1 MGNLVCCVQVDQSQVAMKEGFGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCET 60
MGNL CCVQVDQS VA+KE FG+FE+VLQPGCHC+PW LG ++AGHLS+RVQQLD+KCET
Sbjct: 147 MGNLFCCVQVDQSTVAIKERFGRFEEVLQPGCHCLPWILGSQLAGHLSIRVQQLDVKCET 326
Query: 61 KTKDNVFVNVVASIQYRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQ 120
KTKDNVFVNVVAS+QYRAL++KA+DAFYKLSNT+ QIQAYVFDVIRASVPKL LDD FEQ
Sbjct: 327 KTKDNVFVNVVASVQYRALSEKASDAFYKLSNTKTQIQAYVFDVIRASVPKLLLDDAFEQ 506
Query: 121 KNEIAKAVEEELEKAMSAYGYEIVQTLITDIE 152
KN+IAKAVEEELEKAMSAYGYEIVQTLI DIE
Sbjct: 507 KNDIAKAVEEELEKAMSAYGYEIVQTLIVDIE 602
>TC9262 similar to UP|Q9FHM7 (Q9FHM7) Gb|AAF03497.1 (At5g51570), partial
(63%)
Length = 584
Score = 198 bits (503), Expect = 1e-51
Identities = 102/185 (55%), Positives = 130/185 (70%), Gaps = 2/185 (1%)
Frame = +1
Query: 1 MGNLVC--CVQVDQSQVAMKEGFGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKC 58
MGN C C + Q+ V + E +G+F +V QPG H G+ +AG LS R+ LD++
Sbjct: 28 MGNTFCFFCGCIGQASVGVVEHWGRFHRVAQPGFHFFNPLAGECLAGVLSTRIASLDVRI 207
Query: 59 ETKTKDNVFVNVVASIQYRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTF 118
ETKTKDNVFV ++ SIQYR + + A+DAFY+L N + QIQAYVFDV RA VP++NLDD F
Sbjct: 208 ETKTKDNVFVQLLCSIQYRVVKENADDAFYELQNPQEQIQAYVFDVTRAIVPRMNLDDLF 387
Query: 119 EQKNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKA 178
EQK E+AKAV EEL K M YGY I L+ DI PD V++AMNEINAA RM LA++ K
Sbjct: 388 EQKGEVAKAVLEELHKVMGEYGYSIEHILMVDIIPDPSVRKAMNEINAAQRMLLASEFKG 567
Query: 179 EAEKI 183
+AEK+
Sbjct: 568 DAEKV 582
>AV775598
Length = 448
Score = 125 bits (314), Expect(2) = 3e-30
Identities = 73/134 (54%), Positives = 83/134 (61%), Gaps = 8/134 (5%)
Frame = +3
Query: 29 QPGCHCMP--------WFLGKRIAGHLSLRVQQLDIKCETKTKDNVFVNVVASIQYRALA 80
+PGCHC+P WF Q +C+ + YRAL
Sbjct: 66 RPGCHCVPCMYR*SDFWFPFSACEAA*CS**NQNQGQCDL*LW-------LLQSSYRALT 224
Query: 81 DKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEELEKAMSAYG 140
DKA DA+YKLS+T+ QIQAYVFDVIRA PK+ LD EQKNE+A AVEEELEKAMSAYG
Sbjct: 225 DKAVDAYYKLSDTKTQIQAYVFDVIRAQCPKMELDSAPEQKNEMATAVEEELEKAMSAYG 404
Query: 141 YEIVQTLITDIEPD 154
YEIVQTLI DIEPD
Sbjct: 405 YEIVQTLIVDIEPD 446
Score = 22.7 bits (47), Expect(2) = 3e-30
Identities = 10/12 (83%), Positives = 11/12 (91%)
Frame = +2
Query: 8 VQVDQSQVAMKE 19
VQVDQS VA+KE
Sbjct: 2 VQVDQSSVAIKE 37
>TC17023 homologue to UP|Q9FM19 (Q9FM19) Hypersensitive-induced response
protein, partial (29%)
Length = 510
Score = 101 bits (251), Expect = 2e-22
Identities = 52/65 (80%), Positives = 56/65 (86%)
Frame = +3
Query: 200 GLGIARQRQAIVDGLRDSVIGFSVNVPGTTAKDVMDMVLVTQYFDTMKEIGAASKSSAVF 259
G+GIARQRQAIVDGLRDSV+GFS NVPGTTAKDV+DMVLVTQYFDTMKEIGA S+ F
Sbjct: 6 GVGIARQRQAIVDGLRDSVLGFSGNVPGTTAKDVLDMVLVTQYFDTMKEIGATSQVFCCF 185
Query: 260 IPHGP 264
H P
Sbjct: 186 --HSP 194
Score = 56.6 bits (135), Expect = 5e-09
Identities = 28/36 (77%), Positives = 30/36 (82%)
Frame = +1
Query: 247 KEIGAASKSSAVFIPHGPGAVRDVASQIRDGLLQGS 282
K++ KSSAVFIPHGPGAVRDVAS IRDGLLQ S
Sbjct: 148 KKLVLPPKSSAVFIPHGPGAVRDVASPIRDGLLQAS 255
>TC14853 similar to UP|PZ12_LUPPO (P16148) PPLZ12 protein, partial (40%)
Length = 531
Score = 95.1 bits (235), Expect = 1e-20
Identities = 38/71 (53%), Positives = 61/71 (85%)
Frame = +3
Query: 212 DGLRDSVIGFSVNVPGTTAKDVMDMVLVTQYFDTMKEIGAASKSSAVFIPHGPGAVRDVA 271
DGLR++++ FS + GT+AK+VMD+++VTQYFDT+K++G +SK++ +FIPHGPG VRD+
Sbjct: 3 DGLRENILNFSGKLEGTSAKEVMDLIMVTQYFDTIKDLGNSSKNTTIFIPHGPGHVRDIG 182
Query: 272 SQIRDGLLQGS 282
QIR+G+++ +
Sbjct: 183 DQIRNGVMEAA 215
>TC7841 homologue to UP|1433_OENHO (P29307) 14-3-3-like protein, partial
(12%)
Length = 446
Score = 45.8 bits (107), Expect = 8e-06
Identities = 19/27 (70%), Positives = 23/27 (84%)
Frame = -1
Query: 10 VDQSQVAMKEGFGKFEKVLQPGCHCMP 36
VDQS VA+KE GK++ VLQPGCHC+P
Sbjct: 83 VDQSSVAIKEVNGKYDDVLQPGCHCVP 3
>TC14757 similar to UP|O04361 (O04361) Prohibitin, complete
Length = 1141
Score = 37.7 bits (86), Expect = 0.002
Identities = 26/99 (26%), Positives = 46/99 (46%), Gaps = 3/99 (3%)
Frame = +3
Query: 103 DVIRASVPKLNLDDTFEQKNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMN 162
+V++A V + N D ++ +++ V E L + + + IT + RA+
Sbjct: 423 EVLKAVVAQFNADQLLTERPQVSALVRESLLRRAKDFNILLDDVAITHLSYGAEFSRAVE 602
Query: 163 EINAAARMRLAAK---EKAEAEKILQIKRAEGEAESKYL 198
+ A + +K KAE E+ I RAEGE+E+ L
Sbjct: 603 QKQVAQQEAERSKFVVMKAEQERRAAIIRAEGESEAAKL 719
>TC9327 similar to UP|Q38921 (Q38921) GTP-binding protein ATGB1
(GTP-binding protein-like) (At5g52210), partial (27%)
Length = 538
Score = 26.2 bits (56), Expect = 6.8
Identities = 12/31 (38%), Positives = 16/31 (50%)
Frame = +1
Query: 28 LQPGCHCMPWFLGKRIAGHLSLRVQQLDIKC 58
L+P H M W LGK + G L + +KC
Sbjct: 64 LKPFQHMMGWVLGKVLNGWCKLWTEARGLKC 156
>TC13340 similar to UP|Q94A50 (Q94A50) AT3g55070/T15C9_70, partial (35%)
Length = 466
Score = 25.8 bits (55), Expect = 8.9
Identities = 18/73 (24%), Positives = 29/73 (39%)
Frame = +2
Query: 118 FEQKNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEK 177
FE + +A +EK MSA + D PD A+N +N+ K K
Sbjct: 152 FEHYKKTIRANHRAVEKEMSAVISGVTDAAAADFSPD----DAVNHLNSLVSRLQGLKRK 319
Query: 178 AEAEKILQIKRAE 190
E + + +A+
Sbjct: 320 LEEGSLAEHLQAQ 358
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,443,074
Number of Sequences: 28460
Number of extensions: 33633
Number of successful extensions: 174
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 173
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 174
length of query: 286
length of database: 4,897,600
effective HSP length: 89
effective length of query: 197
effective length of database: 2,364,660
effective search space: 465838020
effective search space used: 465838020
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146776.3


