

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146775.17 + phase: 0 /pseudo
(140 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
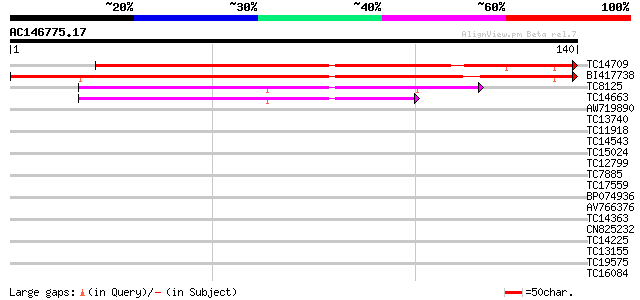
Score E
Sequences producing significant alignments: (bits) Value
TC14709 similar to UP|Q944L1 (Q944L1) At1g70780/F5A18_4, partial... 173 9e-45
BI417738 132 3e-32
TC8125 59 4e-10
TC14663 similar to UP|Q7S2G7 (Q7S2G7) Predicted protein, partial... 52 5e-08
AW719890 28 0.77
TC13740 27 1.3
TC11918 weakly similar to Q7TXG7 (Q7TXG7) Possible conserved tra... 26 2.2
TC14543 similar to UP|Q9ZSY2 (Q9ZSY2) ALTERED response to gravit... 26 2.2
TC15024 homologue to UP|Q9SAU1 (Q9SAU1) TATA-box binding protein... 26 2.2
TC12799 weakly similar to UP|Q9FM10 (Q9FM10) Similarity to MtN3 ... 26 2.2
TC7885 similar to UP|RL4B_ARATH (Q9SF40) 60S ribosomal protein L... 26 2.9
TC17559 weakly similar to UP|Q9LY31 (Q9LY31) Palmitoyl-protein t... 26 2.9
BP074936 25 3.8
AV766376 25 3.8
TC14363 homologue to UP|CAT4_SOYBN (O48561) Catalase 4 , partia... 25 5.0
CN825232 25 6.5
TC14225 similar to UP|Q40478 (Q40478) EREBP-4, partial (5%) 25 6.5
TC13155 homologue to UP|Q9SXT0 (Q9SXT0) Chalcone reductase (Frag... 24 8.5
TC19575 24 8.5
TC16084 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17H... 24 8.5
>TC14709 similar to UP|Q944L1 (Q944L1) At1g70780/F5A18_4, partial (52%)
Length = 690
Score = 173 bits (439), Expect = 9e-45
Identities = 90/122 (73%), Positives = 102/122 (82%), Gaps = 3/122 (2%)
Frame = +1
Query: 22 SINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGNDHSAFFLYCPHLGSDAA 81
SINVLGSAGPIRFV+NEEELV AVIDT LKSYAREGRLPVLG D + F LYCPH+GSD A
Sbjct: 1 SINVLGSAGPIRFVLNEEELVAAVIDTALKSYAREGRLPVLGTDTTGFALYCPHVGSD-A 177
Query: 82 LSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGTSSLP-RRGSGSWKSWFN--LNLKV 138
LSPWD+IGSHGARNF+LCKKPQ + A + +GT+ LP RR SGSWK+WFN L+LK+
Sbjct: 178 LSPWDRIGSHGARNFMLCKKPQGSPGVA---ESAGTTPLPRRRSSGSWKAWFNKSLSLKI 348
Query: 139 SS 140
SS
Sbjct: 349 SS 354
>BI417738
Length = 598
Score = 132 bits (331), Expect = 3e-32
Identities = 76/145 (52%), Positives = 97/145 (66%), Gaps = 5/145 (3%)
Frame = +1
Query: 1 MLLSKQKKNQNANNAK---RRLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREG 57
M+L K + + +++ K R LI+IN+LGSAGPIRFVVNEE+LV VIDT LKSYAREG
Sbjct: 169 MVLQKNRGSVYSDDQKAKTNRFLITINILGSAGPIRFVVNEEKLVSGVIDTALKSYAREG 348
Query: 58 RLPVLGNDHSAFFLYCPHLGSDAALSPWDKIGSHGARNFVLCKKPQAATNEAAAEDGSGT 117
RLPVLG D S F LY + G D AL+P + IGS+G RNFVLCKKP ++ + +
Sbjct: 349 RLPVLGFDASNFLLYRANAGFD-ALNPLEPIGSYGERNFVLCKKPVYHPSKTEPQ----S 513
Query: 118 SSLPRRGSGSWKSWFN--LNLKVSS 140
L ++ SG WK+W N LK+ S
Sbjct: 514 ELLSQKSSGGWKAWLNKSFGLKILS 588
>TC8125
Length = 1373
Score = 58.5 bits (140), Expect = 4e-10
Identities = 36/102 (35%), Positives = 56/102 (54%), Gaps = 2/102 (1%)
Frame = +2
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVL-GNDHSAFFLYCPHL 76
+LL+ + VLGS GP++ ++ E V ++ + Y +E R P+L ND S F L+
Sbjct: 425 KLLLKVTVLGSLGPVQVLMRPESTVGELVAAVARQYVKEARRPILPTNDASDFDLHYSQF 604
Query: 77 GSDAALSPWDKIGSHGARNFVLC-KKPQAATNEAAAEDGSGT 117
+ +L +K+ G+RNF +C +K AAT AA E G T
Sbjct: 605 SLE-SLHREEKLVELGSRNFFMCPRKKLAATTPAAVEGGGVT 727
>TC14663 similar to UP|Q7S2G7 (Q7S2G7) Predicted protein, partial (3%)
Length = 1141
Score = 51.6 bits (122), Expect = 5e-08
Identities = 27/85 (31%), Positives = 48/85 (55%), Gaps = 1/85 (1%)
Frame = +3
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVL-GNDHSAFFLYCPHL 76
+LL+ + ++ S GP+ V++ E V ++ TL+ Y +EGR P+L D S F L+
Sbjct: 477 KLLLKVTMMRSLGPVLMVMSPESTVGDLVAATLRQYVKEGRRPILPSGDPSQFDLHYSQF 656
Query: 77 GSDAALSPWDKIGSHGARNFVLCKK 101
+ +L +K+ G+RNF +C +
Sbjct: 657 SLE-SLDRKEKLSDIGSRNFFMCPR 728
>AW719890
Length = 599
Score = 27.7 bits (60), Expect = 0.77
Identities = 11/22 (50%), Positives = 15/22 (68%)
Frame = +2
Query: 81 ALSPWDKIGSHGARNFVLCKKP 102
+L P +K+ G+RNF LC KP
Sbjct: 461 SLKPDEKVMQLGSRNFFLCSKP 526
>TC13740
Length = 562
Score = 26.9 bits (58), Expect = 1.3
Identities = 13/38 (34%), Positives = 20/38 (52%)
Frame = +1
Query: 101 KPQAATNEAAAEDGSGTSSLPRRGSGSWKSWFNLNLKV 138
+P+ ++ EA EDG R K W+NL+L+V
Sbjct: 1 RPKRSSKEAN-EDGQAREKRSSRDRSKKKKWYNLHLRV 111
>TC11918 weakly similar to Q7TXG7 (Q7TXG7) Possible conserved transmembrane
protein, partial (21%)
Length = 633
Score = 26.2 bits (56), Expect = 2.2
Identities = 13/34 (38%), Positives = 15/34 (43%)
Frame = +3
Query: 63 GNDHSAFFLYCPHLGSDAALSPWDKIGSHGARNF 96
G+DHS F H + LS W SH R F
Sbjct: 129 GSDHSTFSCSGQHPNRGSDLSSWGHDWSHHGRGF 230
>TC14543 similar to UP|Q9ZSY2 (Q9ZSY2) ALTERED response to gravity (ARG1
protein), partial (50%)
Length = 1078
Score = 26.2 bits (56), Expect = 2.2
Identities = 15/40 (37%), Positives = 20/40 (49%), Gaps = 5/40 (12%)
Frame = +1
Query: 105 ATNEAAAEDGSGTSSLPRRGS-----GSWKSWFNLNLKVS 139
++++ + ED G S GS K WFNLNLK S
Sbjct: 580 SSSKISGEDSKGESPGEEGGSDGKDKSGKKKWFNLNLKSS 699
>TC15024 homologue to UP|Q9SAU1 (Q9SAU1) TATA-box binding protein
(Transcription initiation factor TFIID) (TATA-box
factor) (TATA sequence-binding protein) (TBP), partial
(29%)
Length = 1334
Score = 26.2 bits (56), Expect = 2.2
Identities = 20/67 (29%), Positives = 29/67 (42%)
Frame = -2
Query: 5 KQKKNQNANNAKRRLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGN 64
K + Q NN+KR IN ++ ++ +I T +S R+ PVL N
Sbjct: 757 KTYQKQRQNNSKRLS*FYIN------------SDSDVSLVLIFTVSRSSLRDHTAPVLDN 614
Query: 65 DHSAFFL 71
H FFL
Sbjct: 613 YHCWFFL 593
>TC12799 weakly similar to UP|Q9FM10 (Q9FM10) Similarity to MtN3 protein,
partial (35%)
Length = 806
Score = 26.2 bits (56), Expect = 2.2
Identities = 13/31 (41%), Positives = 15/31 (47%)
Frame = +1
Query: 100 KKPQAATNEAAAEDGSGTSSLPRRGSGSWKS 130
KKP TNEAA GT+ P + W S
Sbjct: 541 KKPTTTTNEAAESMEMGTAKPPPFNAN*WNS 633
>TC7885 similar to UP|RL4B_ARATH (Q9SF40) 60S ribosomal protein L4-2 (L1),
complete
Length = 1554
Score = 25.8 bits (55), Expect = 2.9
Identities = 23/93 (24%), Positives = 37/93 (39%), Gaps = 5/93 (5%)
Frame = -3
Query: 50 LKSYAREGRLPVLGNDHSA-----FFLYCPHLGSDAALSPWDKIGSHGARNFVLCKKPQA 104
L+ + GR+PVL SA L D + + + + G V + P+A
Sbjct: 682 LRIFPLPGRMPVLSLAFSASGKAPICLRTLIASLDFSTPSAESLTTRGNSGTVSMRWPRA 503
Query: 105 ATNEAAAEDGSGTSSLPRRGSGSWKSWFNLNLK 137
T+E AE + +W+ WF L L+
Sbjct: 502 RTSEGTAEAAMAETR-------AWRFWFTLILR 425
>TC17559 weakly similar to UP|Q9LY31 (Q9LY31) Palmitoyl-protein thioesterase
precursor-like, partial (39%)
Length = 574
Score = 25.8 bits (55), Expect = 2.9
Identities = 15/47 (31%), Positives = 21/47 (43%)
Frame = +3
Query: 86 DKIGSHGARNFVLCKKPQAATNEAAAEDGSGTSSLPRRGSGSWKSWF 132
D+ +HG + F T E + G+ + GSGSW SWF
Sbjct: 138 DQCSNHGVKRF---------TEELSTYSGAKGYCI-EIGSGSWDSWF 248
>BP074936
Length = 161
Score = 25.4 bits (54), Expect = 3.8
Identities = 12/23 (52%), Positives = 15/23 (65%)
Frame = -2
Query: 103 QAATNEAAAEDGSGTSSLPRRGS 125
+ A EA AE+G G SS+ RGS
Sbjct: 82 EGAEAEAEAEEGRGCSSMTLRGS 14
>AV766376
Length = 335
Score = 25.4 bits (54), Expect = 3.8
Identities = 13/29 (44%), Positives = 18/29 (61%)
Frame = -1
Query: 111 AEDGSGTSSLPRRGSGSWKSWFNLNLKVS 139
A+ SGTSSL + G G +S F L ++ S
Sbjct: 188 AKISSGTSSLMQNGLGGGQSSFKLRIQFS 102
>TC14363 homologue to UP|CAT4_SOYBN (O48561) Catalase 4 , partial (63%)
Length = 1045
Score = 25.0 bits (53), Expect = 5.0
Identities = 13/40 (32%), Positives = 21/40 (52%), Gaps = 3/40 (7%)
Frame = +3
Query: 35 VVNEEELVEAVIDT---TLKSYAREGRLPVLGNDHSAFFL 71
V++E E + D +K Y REG ++GN+ FF+
Sbjct: 432 VIHERGSPETLRDPRGFAVKFYTREGNFDLVGNNFPVFFV 551
>CN825232
Length = 162
Score = 24.6 bits (52), Expect = 6.5
Identities = 13/27 (48%), Positives = 13/27 (48%)
Frame = +1
Query: 102 PQAATNEAAAEDGSGTSSLPRRGSGSW 128
P AA N A G TSS PRR W
Sbjct: 82 PPAALNAPARLMGRTTSSSPRRSFWGW 162
>TC14225 similar to UP|Q40478 (Q40478) EREBP-4, partial (5%)
Length = 604
Score = 24.6 bits (52), Expect = 6.5
Identities = 9/20 (45%), Positives = 15/20 (75%)
Frame = -1
Query: 51 KSYAREGRLPVLGNDHSAFF 70
++ A E P+LG++HS+FF
Sbjct: 112 QTVAAETETPLLGSEHSSFF 53
>TC13155 homologue to UP|Q9SXT0 (Q9SXT0) Chalcone reductase (Fragment),
partial (76%)
Length = 392
Score = 24.3 bits (51), Expect = 8.5
Identities = 13/31 (41%), Positives = 16/31 (50%)
Frame = -3
Query: 99 CKKPQAATNEAAAEDGSGTSSLPRRGSGSWK 129
C PQ+ TN+ +A G SS P SG K
Sbjct: 111 CNHPQSMTNQRSASSG---SSSPCHSSGPQK 28
>TC19575
Length = 500
Score = 24.3 bits (51), Expect = 8.5
Identities = 10/13 (76%), Positives = 12/13 (91%)
Frame = -1
Query: 114 GSGTSSLPRRGSG 126
GSGT+S+ RRGSG
Sbjct: 170 GSGTASVNRRGSG 132
>TC16084 similar to GB|AAL84954.1|19310435|AY078954 F17H15.1/F17H15.1
{Arabidopsis thaliana;}, partial (29%)
Length = 889
Score = 24.3 bits (51), Expect = 8.5
Identities = 10/16 (62%), Positives = 11/16 (68%)
Frame = -1
Query: 69 FFLYCPHLGSDAALSP 84
FFL HLG DA +SP
Sbjct: 742 FFLLMGHLGEDAKVSP 695
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,540,459
Number of Sequences: 28460
Number of extensions: 31863
Number of successful extensions: 146
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 141
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 142
length of query: 140
length of database: 4,897,600
effective HSP length: 81
effective length of query: 59
effective length of database: 2,592,340
effective search space: 152948060
effective search space used: 152948060
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 50 (23.9 bits)
Medicago: description of AC146775.17


