

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146760.4 + phase: 0
(303 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
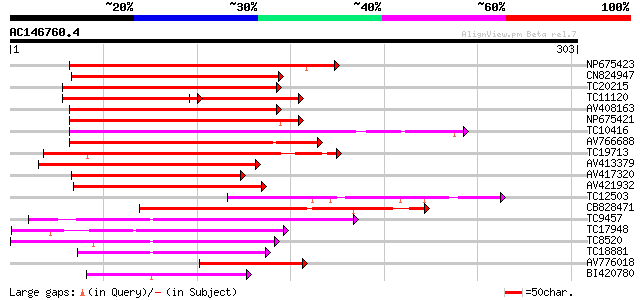
Score E
Sequences producing significant alignments: (bits) Value
NP675423 transcription factor MYB101 [Lotus japonicus] 203 3e-53
CN824947 186 4e-48
TC20215 similar to AAS10104 (AAS10104) MYB transcription factor,... 184 2e-47
TC11120 UP|Q84PP4 (Q84PP4) Transcription factor MYB102, complete 113 3e-46
AV408163 179 5e-46
NP675421 transcription factor MYB103 [Lotus japonicus] 175 7e-45
TC10416 weakly similar to UP|O04108 (O04108) MYB factor, partial... 161 1e-40
AV766688 159 4e-40
TC19713 homologue to UP|P93474 (P93474) Myb26, partial (55%) 157 2e-39
AV413379 157 2e-39
AV417320 152 7e-38
AV421932 145 6e-36
TC12503 homologue to UP|Q9ATD9 (Q9ATD9) BNLGHi233, partial (24%) 142 7e-35
CB828471 134 2e-32
TC9457 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucros... 114 3e-26
TC17948 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose... 110 2e-25
TC8520 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucros... 108 1e-24
TC18881 similar to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucrose... 97 4e-21
AV776018 81 2e-16
BI420780 77 5e-15
>NP675423 transcription factor MYB101 [Lotus japonicus]
Length = 1047
Score = 203 bits (516), Expect = 3e-53
Identities = 91/148 (61%), Positives = 110/148 (73%), Gaps = 4/148 (2%)
Frame = +1
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
TPCC ++GLK+GPWT EED +L +YI+K G G WR LPK AGL RCGKSCRLRW NYLRP
Sbjct: 10 TPCCDEIGLKKGPWTPEEDAILVDYIQKHGHGSWRALPKLAGLNRCGKSCRLRWTNYLRP 189
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG+ +EE LI+ LH +LGN+WS IAG +PGRTDNEIKN+WNTHL KKL+ G+DP
Sbjct: 190 DIKRGKFTEEEEQLIINLHAVLGNKWSAIAGHLPGRTDNEIKNFWNTHLKKKLLQMGLDP 369
Query: 153 RTHKP----LNNPSSSSSIIPNTNHQTS 176
TH+P LN S+ + TN TS
Sbjct: 370 VTHRPRLDHLNLLSNLQQFLAATNMVTS 453
>CN824947
Length = 738
Score = 186 bits (472), Expect = 4e-48
Identities = 81/113 (71%), Positives = 96/113 (84%)
Frame = +1
Query: 34 PCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPS 93
PCC KVGLKRG WT EEDE+L++YI+ GEG WR+LPK AGLLRCGKSCRLRW+NYLR
Sbjct: 163 PCCEKVGLKRGRWTAEEDELLTKYIQASGEGSWRSLPKNAGLLRCGKSCRLRWINYLRGD 342
Query: 94 VKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLI 146
+KRG I+ +EE LI++LH GNRWSLIA +PGRTDNEIKNYWN+HLS+KL+
Sbjct: 343 LKRGNISDEEESLIVKLHASFGNRWSLIASHMPGRTDNEIKNYWNSHLSRKLV 501
>TC20215 similar to AAS10104 (AAS10104) MYB transcription factor, partial
(35%)
Length = 464
Score = 184 bits (467), Expect = 2e-47
Identities = 78/117 (66%), Positives = 98/117 (83%)
Frame = +2
Query: 29 TTTTTPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMN 88
T PCC KVGLKRG W+ EED++L+++IK+ GEG W++LPK AGLLRCGKSCRLRW+N
Sbjct: 26 TMGRAPCCEKVGLKRGRWSEEEDKILTDFIKQNGEGSWKSLPKNAGLLRCGKSCRLRWIN 205
Query: 89 YLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKL 145
YLR VKRG I +EE++I++LH LGNRWS+IAG +PGRTDNEIKNYWN+HL +K+
Sbjct: 206 YLREDVKRGNITSEEEEIIVKLHAALGNRWSVIAGHLPGRTDNEIKNYWNSHLRRKI 376
>TC11120 UP|Q84PP4 (Q84PP4) Transcription factor MYB102, complete
Length = 1281
Score = 113 bits (282), Expect(2) = 3e-46
Identities = 48/75 (64%), Positives = 58/75 (77%)
Frame = +1
Query: 29 TTTTTPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMN 88
T +PCC + GLK+GPWT EED+ L E+I+K G G WR LPK AGL RCGKSCRLRW N
Sbjct: 64 TMGRSPCCDENGLKKGPWTPEEDQKLVEHIQKHGHGSWRALPKLAGLNRCGKSCRLRWTN 243
Query: 89 YLRPSVKRGQIAPDE 103
YLRP +KRG+ +P+E
Sbjct: 244 YLRPDIKRGKFSPEE 288
Score = 88.6 bits (218), Expect(2) = 3e-46
Identities = 38/61 (62%), Positives = 44/61 (71%)
Frame = +2
Query: 97 GQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDPRTHK 156
G +E IL LH +LGN+WS IA +PGRTDNEIKN+WNTHL KKLI G DP TH+
Sbjct: 269 GNFLQKKEQTILHLHSILGNKWSTIATHLPGRTDNEIKNFWNTHLKKKLIQMGFDPMTHQ 448
Query: 157 P 157
P
Sbjct: 449 P 451
>AV408163
Length = 425
Score = 179 bits (454), Expect = 5e-46
Identities = 76/113 (67%), Positives = 93/113 (82%)
Frame = +2
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
+PCC K + +G WT EED+ L YI+ GEG WR+LPK AGLLRCGKSCRLRW+NYLRP
Sbjct: 86 SPCCEKAHINKGAWTKEEDDRLISYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRP 265
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKL 145
+KRG +E++LI++LH LLGN+WSLIAGR+PGRTDNEIKNYWNTH+ +KL
Sbjct: 266 DLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKL 424
>NP675421 transcription factor MYB103 [Lotus japonicus]
Length = 924
Score = 175 bits (444), Expect = 7e-45
Identities = 78/127 (61%), Positives = 96/127 (75%), Gaps = 2/127 (1%)
Frame = +1
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
TP + G+K+GPW+ EED+VL +YI+K G G WR LP+ AGL RCGKSCRLRW NYLRP
Sbjct: 10 TPSIDESGMKKGPWSQEEDKVLVDYIQKHGHGSWRALPQLAGLNRCGKSCRLRWTNYLRP 189
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSK--KLINQGI 150
+KRG+ + +EE LI+ LH LGN+W+ IA +PGRTDNEIKN WNTHL K KL+ G+
Sbjct: 190 DIKRGKFSDEEEQLIINLHASLGNKWATIASHLPGRTDNEIKNLWNTHLKKKLKLLQMGL 369
Query: 151 DPRTHKP 157
DP TH P
Sbjct: 370 DPVTHCP 390
>TC10416 weakly similar to UP|O04108 (O04108) MYB factor, partial (41%)
Length = 1046
Score = 161 bits (407), Expect = 1e-40
Identities = 82/216 (37%), Positives = 122/216 (55%), Gaps = 3/216 (1%)
Frame = +2
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
TP C K G+++G WT EED+ L Y+ + G WR LPK AGL RCGKSCRLRW+NYLRP
Sbjct: 101 TPSCDKSGMRKGTWTAEEDKKLIAYVTRYGCWNWRQLPKFAGLARCGKSCRLRWLNYLRP 280
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
++KRG +EE++I+R+H+ LGNRWS IA +PGRTD+E+KN+W+T L + + Q
Sbjct: 281 NIKRGNFTQEEEEIIIRMHKKLGNRWSTIAAELPGRTDSEVKNHWHTTLKRTVHEQNTVT 460
Query: 153 RTHKPLNNPSSSSSIIPNTNHQTSTPFFAPSSSDHHHPITITNNEPDPSQTQQAQGCGNL 212
++N + +P + + P++S I+ + P T C
Sbjct: 461 NEETKVSNSKNERDPVPKSGIASLLVTPPPATS-----ISGSIGALSPFSTSSEFSC--T 619
Query: 213 VNDVSAMDNHHVLTNNNRGDYGDD---NGNNNYGGD 245
+D++ + +L + D+ DD N N N+ D
Sbjct: 620 TSDLTTSSSSEMLVFEDDFDFLDDFTENVNENFWSD 727
>AV766688
Length = 608
Score = 159 bits (403), Expect = 4e-40
Identities = 72/136 (52%), Positives = 95/136 (68%), Gaps = 1/136 (0%)
Frame = +2
Query: 33 TPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRP 92
TP C K GL++G WT EED L Y+ + G WR LPK AGL RCGKSCRLRW+NYLRP
Sbjct: 86 TPSCDKSGLRKGTWTAEEDRKLIAYVTRYGCWNWRQLPKFAGLARCGKSCRLRWLNYLRP 265
Query: 93 SVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+KRG +EE++I+R+H+ LGNRWS+IA +PGRTDNE+KN+W+T L K+ + Q +
Sbjct: 266 DIKRGHFTQEEEEIIIRMHKNLGNRWSIIAAELPGRTDNEVKNHWHTSL-KRRVQQNEEA 442
Query: 153 RTHKPLN-NPSSSSSI 167
+ N P+S + I
Sbjct: 443 KVSNSKNEGPTSGNGI 490
>TC19713 homologue to UP|P93474 (P93474) Myb26, partial (55%)
Length = 532
Score = 157 bits (398), Expect = 2e-39
Identities = 81/161 (50%), Positives = 102/161 (63%), Gaps = 2/161 (1%)
Frame = +3
Query: 19 SNSNPNKKQKTTTTTPCCSKVG--LKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLL 76
S+ + ++ KT PC S +++GPWT+EED +L YI GEG W TL K AGL
Sbjct: 33 SSVSLKRQHKTMDKKPCNSSHDPEVRKGPWTIEEDLILINYIANHGEGVWNTLAKSAGLK 212
Query: 77 RCGKSCRLRWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNY 136
R GKSCRLRW+NYLRP V+RG I P+E+ LI+ LH GNRWS IA +PGRTDNEIKN+
Sbjct: 213 RTGKSCRLRWLNYLRPDVRRGNITPEEQLLIMELHAKWGNRWSKIAKHLPGRTDNEIKNF 392
Query: 137 WNTHLSKKLINQGIDPRTHKPLNNPSSSSSIIPNTNHQTST 177
W T + K + + + L SSS I N +HQTST
Sbjct: 393 WRTRIQKHI-------KQAESLQQQQSSSEI--NDHHQTST 488
>AV413379
Length = 416
Score = 157 bits (397), Expect = 2e-39
Identities = 70/119 (58%), Positives = 86/119 (71%)
Frame = +3
Query: 16 NSNSNSNPNKKQKTTTTTPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGL 75
N+ + +K +PCC K +G WT EED+ L YI+ GEG WR+LPK AGL
Sbjct: 60 NTTQHKEEEEKLINMGRSPCCEKAHTNKGAWTKEEDDRLIAYIRAHGEGCWRSLPKSAGL 239
Query: 76 LRCGKSCRLRWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIK 134
LRCGKSCRLRW+NYLRP +KRG P E++LI++LH LLGN+WSLIAGR+ GRTDNEIK
Sbjct: 240 LRCGKSCRLRWINYLRPDLKRGNFTPQEDELIIKLHSLLGNKWSLIAGRLAGRTDNEIK 416
>AV417320
Length = 369
Score = 152 bits (384), Expect = 7e-38
Identities = 62/93 (66%), Positives = 76/93 (81%)
Frame = +3
Query: 34 PCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPS 93
PCC K+GLK+GPWT EED++L YI+K G G WR LPK AGLLRCGKSCRLRW+NYLRP
Sbjct: 90 PCCEKIGLKKGPWTSEEDQILISYIQKHGHGNWRALPKHAGLLRCGKSCRLRWINYLRPD 269
Query: 94 VKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIP 126
+KRG +EE+LI+++H LLGNRWS IA ++P
Sbjct: 270 IKRGNFTAEEEELIIKMHELLGNRWSAIAAKLP 368
>AV421932
Length = 402
Score = 145 bits (367), Expect = 6e-36
Identities = 62/103 (60%), Positives = 79/103 (76%)
Frame = +3
Query: 35 CCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPSV 94
CC++ +KRG W+ EEDE L YI G G W +P++AGL RCGKSCRLRW+NYLRP +
Sbjct: 93 CCNQQKVKRGLWSPEEDEKLIRYITTHGYGCWSEVPEKAGLQRCGKSCRLRWINYLRPDI 272
Query: 95 KRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYW 137
+RG+ P+EE LI+ LH ++GNRW+ IA +PGRTDNEIKNYW
Sbjct: 273 RRGRFTPEEEKLIISLHGVVGNRWAHIASHLPGRTDNEIKNYW 401
>TC12503 homologue to UP|Q9ATD9 (Q9ATD9) BNLGHi233, partial (24%)
Length = 957
Score = 142 bits (358), Expect = 7e-35
Identities = 87/172 (50%), Positives = 102/172 (58%), Gaps = 23/172 (13%)
Frame = +2
Query: 117 RWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDPRTHKPLNN----PSSSSSIIPN-- 170
RWSLIAGRIPGRTDNEIKN+WNTHLSKKLI+QGIDPRTHKPLN PSSS++ P
Sbjct: 152 RWSLIAGRIPGRTDNEIKNFWNTHLSKKLISQGIDPRTHKPLNPELSIPSSSTTPPPPPP 331
Query: 171 -------TNHQTSTPFFAPSSSDHHHPITITNNEPDPSQTQQAQ------GCGNLVNDVS 217
TNHQT +++ HH I N E D Q Q Q + ND+
Sbjct: 332 SKPLPVITNHQT----LHETTTHHHLNIASVNQECDEGQYQYQQPQAVAAAAYMVANDIP 499
Query: 218 AMD----NHHVLTNNNRGDYGDDNGNNNYGGDDVFTSFLDSLINDDAFAAHR 265
D H + NNN DY D NY DD+F+SFL+SLIN+DAF+A R
Sbjct: 500 DDDVPSMGHLLANNNNNQDYND----INYWSDDIFSSFLNSLINEDAFSAQR 643
>CB828471
Length = 525
Score = 134 bits (337), Expect = 2e-32
Identities = 75/158 (47%), Positives = 97/158 (60%), Gaps = 3/158 (1%)
Frame = +1
Query: 70 PKRAGLLRCGKSCRLRWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRT 129
PK+AGLLRCGKSCRLRW NYLRP +KRG + +EED I+ LH LLGNRWS IA R+PGRT
Sbjct: 1 PKQAGLLRCGKSCRLRWANYLRPDIKRGNFSREEEDEIINLHELLGNRWSAIAARLPGRT 180
Query: 130 DNEIKNYWNTHLSKKLINQGIDPRTHKPLNNPSSSSSIIPNTNHQTSTPFFAP---SSSD 186
DNEIKN W+THL K+L Q +T+ L+ +S S+ HQ +P SSS+
Sbjct: 181 DNEIKNVWHTHLKKRLQPQTKQKQTNLDLD--ASKSNKDAKKEHQEDEGPRSPQQCSSSN 354
Query: 187 HHHPITITNNEPDPSQTQQAQGCGNLVNDVSAMDNHHV 224
+ ++TN+ T +D+S D H+V
Sbjct: 355 SDNMSSLTNSSSIAPSTNH--------DDMSINDVHNV 444
>TC9457 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and
sucrose-responsive element binding factor, partial (43%)
Length = 860
Score = 114 bits (284), Expect = 3e-26
Identities = 66/178 (37%), Positives = 92/178 (51%), Gaps = 2/178 (1%)
Frame = +3
Query: 11 KSETTNSNSNSNPNKKQKTTTTTPCCSKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLP 70
KS T N+N+N N +K+ +GPW+ EEDE L ++K G W +
Sbjct: 9 KSLVTKMNTNTNTNTS---------ITKMDRIKGPWSPEEDEALQRLVEKHGPRNWSLIS 161
Query: 71 KRAGLLRCGKSCRLRWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTD 130
K R GKSCRLRW N L P V+ +E+D I+R H GN+W+ IA + GRTD
Sbjct: 162 KSIPG-RSGKSCRLRWCNQLSPQVEHRAFTAEEDDTIIRAHARFGNKWATIARLLSGRTD 338
Query: 131 NEIKNYWNTHLSKKLINQGIDPRTHKPLNNPSSSSSIIP-NTNHQTSTPF-FAPSSSD 186
N IKN+WN+ L +K + +D PL S+ + P +T + P +PS SD
Sbjct: 339 NAIKNHWNSTLKRKCASFMMDDHNAPPLKRSVSAGAATPVSTGLYMNPPSPGSPSGSD 512
>TC17948 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose-responsive
element binding factor, partial (50%)
Length = 487
Score = 110 bits (276), Expect = 2e-25
Identities = 57/151 (37%), Positives = 87/151 (56%), Gaps = 3/151 (1%)
Frame = +2
Query: 2 NLSSSTDQDKSETTNSNSN---SNPNKKQKTTTTTPCCSKVGLKRGPWTVEEDEVLSEYI 58
N SSST S++ +S+SN +NPN+ ++ +GPW+ EED +L+ +
Sbjct: 77 NRSSSTTTSSSDSCSSDSNPRSNNPNRPERI-------------KGPWSAEEDRILTRLV 217
Query: 59 KKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPSVKRGQIAPDEEDLILRLHRLLGNRW 118
++ G W +L R R GKSCRLRW N L P+V+ + E++ I+ H GNRW
Sbjct: 218 ERHGARNW-SLISRYIKGRSGKSCRLRWCNQLSPTVEHRPFSVQEDETIIAAHARFGNRW 394
Query: 119 SLIAGRIPGRTDNEIKNYWNTHLSKKLINQG 149
+ IA +PGRTDN +KN+WN+ L +++ G
Sbjct: 395 AAIARLLPGRTDNAVKNHWNSTLKRRVPGGG 487
>TC8520 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and
sucrose-responsive element binding factor, partial (36%)
Length = 1778
Score = 108 bits (269), Expect = 1e-24
Identities = 58/152 (38%), Positives = 80/152 (52%), Gaps = 8/152 (5%)
Frame = +1
Query: 1 MNLSSS-TDQDKSETTNSNSNSNPNKKQKTTTTTPCCSKVGLKR-------GPWTVEEDE 52
+N+S+S T K T S+ KK S + + R GPW+ EEDE
Sbjct: 34 LNISNSHTHTSKLNTLTSHQQQQQQKKSYKKIVQHTYSVMAMTRKEMDRIKGPWSPEEDE 213
Query: 53 VLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPSVKRGQIAPDEEDLILRLHR 112
L + +++ G W + K R GKSCRLRW N L P V+ P+E+D I+R H
Sbjct: 214 ALQKLVERHGPRNWSLISKSIPG-RSGKSCRLRWCNQLSPQVEHRAFTPEEDDTIIRAHA 390
Query: 113 LLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKK 144
GN+W+ IA + GRTDN IKN+WN+ L +K
Sbjct: 391 RFGNKWATIARLLSGRTDNAIKNHWNSTLKRK 486
Score = 34.7 bits (78), Expect = 0.020
Identities = 18/58 (31%), Positives = 32/58 (55%), Gaps = 1/58 (1%)
Frame = +1
Query: 96 RGQIAPDEEDLILRLHRLLGNR-WSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDP 152
+G +P+E++ + +L G R WSLI+ IPGR+ + W LS ++ ++ P
Sbjct: 184 KGPWSPEEDEALQKLVERHGPRNWSLISKSIPGRSGKSCRLRWCNQLSPQVEHRAFTP 357
>TC18881 similar to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucrose-responsive
element binding factor, partial (28%)
Length = 346
Score = 96.7 bits (239), Expect = 4e-21
Identities = 44/103 (42%), Positives = 62/103 (59%)
Frame = +1
Query: 37 SKVGLKRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPSVKR 96
S++ +GPW+ EEDE L+ ++ G W + K R GKSCRLRW N L P V+
Sbjct: 37 SQMDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPG-RSGKSCRLRWCNQLSPEVEH 213
Query: 97 GQIAPDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNT 139
P+E+ I+R H GN+W+ IA + GRTDN +KN+WN+
Sbjct: 214 RPFTPEEDSTIIRAHARCGNKWATIARILNGRTDNAVKNHWNS 342
>AV776018
Length = 476
Score = 80.9 bits (198), Expect = 2e-16
Identities = 37/58 (63%), Positives = 46/58 (78%)
Frame = +1
Query: 102 DEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLINQGIDPRTHKPLN 159
+E++LI+ LH +LGNRWS A + PGRTDNEIKN WN+ L KKL +GIDP THKPL+
Sbjct: 22 EEDNLIIELHAVLGNRWSQDATQWPGRTDNEIKNLWNSCLKKKLRQRGIDPVTHKPLS 195
>BI420780
Length = 475
Score = 76.6 bits (187), Expect = 5e-15
Identities = 35/90 (38%), Positives = 53/90 (58%), Gaps = 2/90 (2%)
Frame = +1
Query: 42 KRGPWTVEEDEVLSEYIKKEGEGRWRTLPKRAG--LLRCGKSCRLRWMNYLRPSVKRGQI 99
+R W+ EED +L Y+++ G W + +R L R KSC RW NYL+P +K+G +
Sbjct: 199 ERQRWSSEEDALLHAYVQQYGPREWNLVSQRMNTPLNRDTKSCLERWKNYLKPGIKKGSL 378
Query: 100 APDEEDLILRLHRLLGNRWSLIAGRIPGRT 129
+E+ L++ L GN+W IA +PGRT
Sbjct: 379 TKEEQRLVILLQANYGNKWKKIAAEVPGRT 468
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.311 0.129 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,848,734
Number of Sequences: 28460
Number of extensions: 118947
Number of successful extensions: 936
Number of sequences better than 10.0: 136
Number of HSP's better than 10.0 without gapping: 880
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 905
length of query: 303
length of database: 4,897,600
effective HSP length: 90
effective length of query: 213
effective length of database: 2,336,200
effective search space: 497610600
effective search space used: 497610600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146760.4


