

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146759.6 - phase: 0
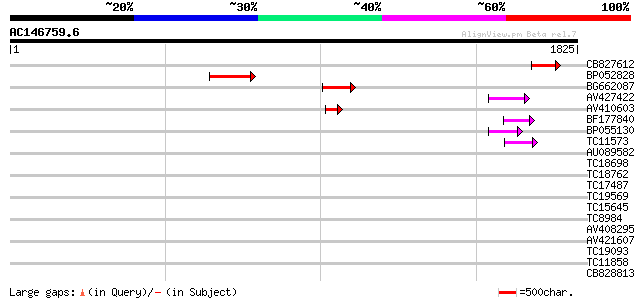
(1825 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB827612 129 5e-30
BP052828 123 2e-28
BG662087 93 4e-19
AV427422 69 5e-12
AV410603 55 1e-07
BF177840 53 4e-07
BP055130 52 6e-07
TC11573 similar to UP|Q9M6N4 (Q9M6N4) Pol protein integrase regi... 47 2e-05
AU089582 40 0.003
TC18698 37 0.037
TC18762 similar to UP|Q9VVG2 (Q9VVG2) CG13731-PA, partial (3%) 36 0.062
TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like prot... 34 0.24
TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor k... 33 0.40
TC15645 similar to UP|ROA3_HUMAN (P51991) Heterogeneous nuclear ... 33 0.53
TC8984 similar to UP|Q8LBA3 (Q8LBA3) Phosphoserine aminotransfer... 32 0.90
AV408295 32 1.2
AV421607 31 2.0
TC19093 similar to UP|P87519 (P87519) GP80, partial (9%) 30 2.6
TC11858 similar to UP|Q9FUQ7 (Q9FUQ7) Calcineurin B-like protein... 30 3.4
CB828813 30 4.5
>CB827612
Length = 277
Score = 129 bits (323), Expect = 5e-30
Identities = 60/92 (65%), Positives = 75/92 (81%)
Frame = +1
Query: 1681 ACHLPVELEHKAYWAIKALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHD 1740
ACHLPVE+EH+AYWA+K+ N + AG +R L+L +LEE+RLGAYE++ IYKE+TK HD
Sbjct: 1 ACHLPVEIEHRAYWAVKSCNXEMQQAGIERKLQLQQLEELRLGAYESSRIYKEKTKHXHD 180
Query: 1741 KGLVRREFYVGQLVLLFNSRLKLFPGKLKSKW 1772
K + R+EF VGQ VLLFNSRLKL GKL+SKW
Sbjct: 181 KMISRKEFSVGQQVLLFNSRLKLMAGKLRSKW 276
>BP052828
Length = 523
Score = 123 bits (309), Expect = 2e-28
Identities = 68/152 (44%), Positives = 94/152 (61%), Gaps = 3/152 (1%)
Frame = +2
Query: 643 AFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLGGGEPKPTRMTLTLADRSISYPFGV 702
A PC+IG + DLGA IN+MP S+ L G + T + + LA+RS + P GV
Sbjct: 59 ALCYPCTIGDNKYENCMLDLGAGINVMPTSIYNILHPGPLQHTGLIVQLANRSNARPKGV 238
Query: 703 LEDVLVKVNDLIFPADFVILDM---AEDEDMPLILGRPFLATGRALIDVEMGQLMLRFHN 759
+EDVLV+VN+LIFPADF ILDM + P+ILGRPF+ T + IDV+ G + + F +
Sbjct: 239 VEDVLVQVNELIFPADFYILDMEGETKSSRAPIILGRPFMKTAKTKIDVDNGTMSMEFGD 418
Query: 760 EQVVFNIFEAMKHRAENPKCYRIDVIDEIVED 791
FNI +AMKH E+ + I V+ E+V+D
Sbjct: 419 IIAKFNIDDAMKHPLEDHSVFHIGVVSELVDD 514
>BG662087
Length = 373
Score = 92.8 bits (229), Expect = 4e-19
Identities = 43/108 (39%), Positives = 67/108 (61%)
Frame = +1
Query: 1006 RMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAF 1065
RM +DY LN A KD +PLP +D++++ + +D YSGY+QI + P D++KTAF
Sbjct: 25 RMWVDYTDLNKACPKDSYPLPSIDKLVDGASDNELLSLMDAYSGYHQIKMHPSDEDKTAF 204
Query: 1066 TCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSV 1113
+ Y+ +PFGL A AT+Q M +F D + +N+EV++D+ V
Sbjct: 205 MTARVNYCYQTIPFGLKNAGATYQXLMDRVFXDXVGRNMEVYLDNMIV 348
>AV427422
Length = 417
Score = 69.3 bits (168), Expect = 5e-12
Identities = 39/135 (28%), Positives = 66/135 (48%), Gaps = 1/135 (0%)
Frame = +1
Query: 1540 PPSSSYLHILVCVDYVTKWVEAIPCV-ANDSKTVVNFLRKNIFTRFGTPRVLISDGGKHF 1598
P S Y +LV VD ++K+ +P +K + + + + G P ++SD F
Sbjct: 13 PKSKGYEAVLVVVDRLSKFSHFVPLKHPYTAKVIADIFVREVVRLHGVPLSIVSDRDPLF 192
Query: 1599 CNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSRKLDDAL 1658
+NF + + K K K++T YHP++ GQ EV NR L+ L +A K W+ + A
Sbjct: 193 MSNFWKELFKMQGTKLKMSTAYHPESDGQTEVVNRCLETYLRCFIADQPKSWAHWVPWAE 372
Query: 1659 WAYRIAFKTHLGLSP 1673
+ Y ++ G +P
Sbjct: 373 YWYNTSYHVSTGQTP 417
>AV410603
Length = 162
Score = 54.7 bits (130), Expect = 1e-07
Identities = 25/54 (46%), Positives = 35/54 (64%)
Frame = +1
Query: 1017 ATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAFTCPFG 1070
+T KD FP+P +D++++ L G F+ LD SGY+QI V PED+ KT F G
Sbjct: 1 STVKDSFPMPTVDELLDELRGSQFFSKLDLRSGYHQILVKPEDRHKTVFRTHHG 162
>BF177840
Length = 410
Score = 53.1 bits (126), Expect = 4e-07
Identities = 30/99 (30%), Positives = 48/99 (48%)
Frame = +2
Query: 1590 LISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKD 1649
++SD F ++F T+ K K +T HPQT GQ EV N+ L +L + + K
Sbjct: 17 IVSDRDTKFISHFWRTLWGKVGTKLLYSTTCHPQTDGQTEVVNKTLSTLLRSVLERNLKM 196
Query: 1650 WSRKLDDALWAYRIAFKTHLGLSPYQLVFGKACHLPVEL 1688
W L +AY + SP+++V+G P++L
Sbjct: 197 WETWLPHIEFAYNRVVHSTTKHSPFEIVYGYNPLTPLDL 313
>BP055130
Length = 567
Score = 52.4 bits (124), Expect = 6e-07
Identities = 34/111 (30%), Positives = 51/111 (45%), Gaps = 1/111 (0%)
Frame = +2
Query: 1541 PSSSYLHILVCVDYVTKWVEAIPCVAN-DSKTVVNFLRKNIFTRFGTPRVLISDGGKHFC 1599
P + I+V VD ++K+ P AN S V I G PR ++SD K F
Sbjct: 170 PVCGFTVIIVVVDRLSKYGHFAPHRANYTSSQVAETFVSTIVKLHGMPRAIVSDRDKAFT 349
Query: 1600 NNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDW 1650
+ F + K + +++ YHPQT GQ E N+ L+ L V + + W
Sbjct: 350 SAFWKHFFKLHGTTLNMSSSYHPQTDGQTEALNKCLELYLRCFVHETPRLW 502
>TC11573 similar to UP|Q9M6N4 (Q9M6N4) Pol protein integrase region
(Fragment), partial (10%)
Length = 572
Score = 47.4 bits (111), Expect = 2e-05
Identities = 26/106 (24%), Positives = 54/106 (50%)
Frame = +2
Query: 1593 DGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSR 1652
+ G F + + I+ + ++ HPQT+GQ E +N+ + + +++ + + W
Sbjct: 8 ENGIQFTSKQTQDFCDGMGIQMRFSSVKHPQTNGQTEAANKVILKGIKRRLYEAEGRWID 187
Query: 1653 KLDDALWAYRIAFKTHLGLSPYQLVFGKACHLPVELEHKAYWAIKA 1698
+L LW+Y ++ + +P+ L +G LPVE+ + + W KA
Sbjct: 188 ELPIVLWSYNTMPQSSIKETPF*LTYGADTMLPVEISNFS-W*TKA 322
>AU089582
Length = 383
Score = 40.4 bits (93), Expect = 0.003
Identities = 22/68 (32%), Positives = 35/68 (51%)
Frame = +2
Query: 1580 IFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQIL 1639
I + G P +ISD G F ++F + + K++T +HPQT GQ E + + L+ +L
Sbjct: 44 IVSLHGVPVSIISDRGAQFTSHFWRSFQTALGTRLKMSTAFHPQTDGQSERTIQILEDML 223
Query: 1640 EKTVASSR 1647
V R
Sbjct: 224 RACVXDLR 247
>TC18698
Length = 808
Score = 36.6 bits (83), Expect = 0.037
Identities = 18/42 (42%), Positives = 25/42 (58%)
Frame = -2
Query: 1072 FAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSV 1113
+ Y+ MP GL T+QR M IF I KN+EV+++D V
Sbjct: 771 YYYQVMPLGLKNI*TTYQRLMDKIFHKQI*KNVEVYVEDMIV 646
>TC18762 similar to UP|Q9VVG2 (Q9VVG2) CG13731-PA, partial (3%)
Length = 759
Score = 35.8 bits (81), Expect = 0.062
Identities = 19/59 (32%), Positives = 26/59 (43%)
Frame = +2
Query: 11 EYDPEIEKTARANRKAVRVSKSVPPGTRTPIPDPTLIETETTTPPPVITMEEGDPPPAR 69
+Y P I K + + A R+ P T T P P+ T+T PP + PPP R
Sbjct: 101 KYPPPITKRQQQTKPAARLPPPPLPTTTTAAPSPSPSTTKTKNSPPPPSPTPKPPPPTR 277
>TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like protein, complete
Length = 1794
Score = 33.9 bits (76), Expect = 0.24
Identities = 31/129 (24%), Positives = 55/129 (42%), Gaps = 8/129 (6%)
Frame = +1
Query: 428 SRKMLETQLGQLAKQLAEQNKGGFSGNTQENPKNETCNVIELRSKKVLAPLVPKVPNKVD 487
+R LE +L + L + G F+G+ + PK E + + L D
Sbjct: 1291 NRAKLELRL----RNLEGKELGRFAGSAKGKPKIEAYDKDRKKGAGGLITPAKTYNPSAD 1458
Query: 488 ESVVEVDENIEDEEVVEKESDQGVVENEKKKKTEGEKSE---RLIDED-----SILRKSK 539
+ + ++ DE+ E +D+ + EKKKK E + + L DED +++K K
Sbjct: 1459 AVISQKSDSAMDEDTHEPSADKKKEKKEKKKKKEKNEEKDVPLLADEDGDEEKEVVKKEK 1638
Query: 540 SQILKDGDE 548
+ K+ E
Sbjct: 1639 KKKRKESTE 1665
>TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor kinase 1;1,
partial (16%)
Length = 486
Score = 33.1 bits (74), Expect = 0.40
Identities = 15/35 (42%), Positives = 19/35 (53%)
Frame = +1
Query: 34 PPGTRTPIPDPTLIETETTTPPPVITMEEGDPPPA 68
PP ++ P P+ T TTTPPP T +PP A
Sbjct: 79 PPPSQQPTHSPSTSPTSTTTPPPSPTKVTENPPTA 183
>TC15645 similar to UP|ROA3_HUMAN (P51991) Heterogeneous nuclear
ribonucleoprotein A3 (hnRNP A3) (D10S102), partial (5%)
Length = 938
Score = 32.7 bits (73), Expect = 0.53
Identities = 20/59 (33%), Positives = 27/59 (44%)
Frame = +1
Query: 35 PGTRTPIPDPTLIETETTTPPPVITMEEGDPPPARPKLGDYGLANHRGRLTHVFRPANP 93
P T + IP + T+TPPP ++ PPPA + + N R R F ANP
Sbjct: 205 PSTNSKIPTLSSAPRSTSTPPPTLSTSLPPPPPA--SKSNRSITNQRPR----FSGANP 363
>TC8984 similar to UP|Q8LBA3 (Q8LBA3) Phosphoserine aminotransferase,
partial (25%)
Length = 563
Score = 32.0 bits (71), Expect = 0.90
Identities = 14/32 (43%), Positives = 16/32 (49%)
Frame = +1
Query: 39 TPIPDPTLIETETTTPPPVITMEEGDPPPARP 70
TPI P + TTTPPP PPP+ P
Sbjct: 82 TPISKPPPTTSSTTTPPPPPPPPSAPPPPSNP 177
>AV408295
Length = 426
Score = 31.6 bits (70), Expect = 1.2
Identities = 24/73 (32%), Positives = 35/73 (47%), Gaps = 15/73 (20%)
Frame = +3
Query: 469 LRSKKVLAPLVPKVPN-----KVDE-SVVEVDENIEDEEV---------VEKESDQGVVE 513
L ++ AP PK P+ K DE + +E +E+EE E+E + VVE
Sbjct: 87 LNKHELSAPPPPKAPSPSTEAKKDEVAAAATEEKVEEEEEKKTEEEKKPAEEEKKEEVVE 266
Query: 514 NEKKKKTEGEKSE 526
EKK E +K+E
Sbjct: 267 EEKKPAEEEKKAE 305
>AV421607
Length = 245
Score = 30.8 bits (68), Expect = 2.0
Identities = 21/63 (33%), Positives = 30/63 (47%)
Frame = +1
Query: 8 EDLEYDPEIEKTARANRKAVRVSKSVPPGTRTPIPDPTLIETETTTPPPVITMEEGDPPP 67
+D + D + E+ A +V+ V PIP PTL T TPPP+ + PPP
Sbjct: 13 QDQDRDQDQEQQQVVTSAADQVTGPVIALPPLPIPIPTL----TLTPPPLPILSFLLPPP 180
Query: 68 ARP 70
+ P
Sbjct: 181SNP 189
>TC19093 similar to UP|P87519 (P87519) GP80, partial (9%)
Length = 525
Score = 30.4 bits (67), Expect = 2.6
Identities = 12/33 (36%), Positives = 18/33 (54%)
Frame = +1
Query: 39 TPIPDPTLIETETTTPPPVITMEEGDPPPARPK 71
TP P P + T PP+ ++ +PPP +PK
Sbjct: 343 TPTPKPRKKYPASKTEPPLQHLQRPEPPPTKPK 441
>TC11858 similar to UP|Q9FUQ7 (Q9FUQ7) Calcineurin B-like protein 5
(Calcineurin B-like protein 8), partial (15%)
Length = 458
Score = 30.0 bits (66), Expect = 3.4
Identities = 17/45 (37%), Positives = 26/45 (57%)
Frame = +1
Query: 1087 TFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFDQCLFHLNAVLKR 1131
+F C+ S++I K+I+ MDDF KSFD L ++L+R
Sbjct: 310 SFSNCL----SNVILKDIDTPMDDFVHVWKSFDH*FNRLESILRR 432
>CB828813
Length = 465
Score = 29.6 bits (65), Expect = 4.5
Identities = 16/50 (32%), Positives = 24/50 (48%), Gaps = 10/50 (20%)
Frame = +1
Query: 29 VSKSVPPGTRTPI----------PDPTLIETETTTPPPVITMEEGDPPPA 68
++K P G+ TP P PT T +T+ PP+ T +PPP+
Sbjct: 178 LTKPHPIGSTTPFSALTTPPPPPPPPTPTRTSSTSKPPIPTPPPPNPPPS 327
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,042,438
Number of Sequences: 28460
Number of extensions: 448149
Number of successful extensions: 2845
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 2610
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2800
length of query: 1825
length of database: 4,897,600
effective HSP length: 103
effective length of query: 1722
effective length of database: 1,966,220
effective search space: 3385830840
effective search space used: 3385830840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146759.6


