

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.5 - phase: 0 /pseudo
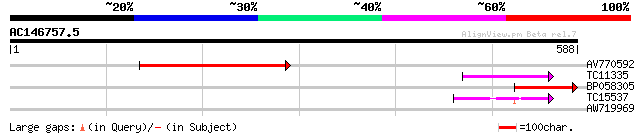
(588 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV770592 216 8e-57
TC11335 89 2e-18
BP058305 84 6e-17
TC15537 52 4e-07
AW719969 27 9.3
>AV770592
Length = 489
Score = 216 bits (550), Expect = 8e-57
Identities = 104/157 (66%), Positives = 122/157 (77%)
Frame = +2
Query: 135 AVVKTTKGFKFGGAIFAEPNCWSMLKGGLIADTTGVAELYFESNNTSVEIWVDNVSLQPF 194
++VKTT+G K GA AE N L+ +T+G AELYFES NTSVEIW+D++SLQPF
Sbjct: 17 SIVKTTEGLKLAGATLAESNVGLCLRVVSQTNTSGTAELYFESENTSVEIWIDSISLQPF 196
Query: 195 TEKQWRSHQELSIEKDRKRKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNN 254
T+KQW SHQ S EK RK KV+V+A++EQG+PLPNASI +T KR GFPFGSAIN NILNN
Sbjct: 197 TQKQWSSHQHQSTEKARKSKVLVQAIDEQGNPLPNASIHITQKRLGFPFGSAINHNILNN 376
Query: 255 NAYQDWFASRFTVTTFENEMKWYTNEYAQGKDNYFDA 291
AYQ+WF SRFTVTTFENEMKWY E QGK NY+ A
Sbjct: 377 GAYQNWFTSRFTVTTFENEMKWYMTETTQGKPNYYYA 487
>TC11335
Length = 447
Score = 89.0 bits (219), Expect = 2e-18
Identities = 41/95 (43%), Positives = 55/95 (57%)
Frame = +2
Query: 470 NQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFKNLPAGDVVDQLINEWG 529
+Q Y + +LRE ++HP +QGI+M+ G + L D NFKN P GDVVD LI EWG
Sbjct: 2 HQEDYVDWILREGYAHPAVQGIIMFMGPVQAGFKKAPLADENFKNTPIGDVVDMLIREWG 181
Query: 530 RAEKSGTTDQNGYFEASLFHGDYEIEINHPIKKKS 564
D G + SL HGDY++ + HP K+ S
Sbjct: 182 TGPHEAKADSRGIVDISLHHGDYDVIVTHPQKQIS 286
>BP058305
Length = 303
Score = 84.0 bits (206), Expect = 6e-17
Identities = 38/66 (57%), Positives = 49/66 (73%), Gaps = 1/66 (1%)
Frame = -3
Query: 524 LINEWGRAEK-SGTTDQNGYFEASLFHGDYEIEINHPIKKKSNFTHHIKVLSKDEFKKTK 582
L++EW ++ SGTTDQNG E SLFHGDYE+EI+HP KK THH++V+ DE K+
Sbjct: 301 LLHEWXXSKTLSGTTDQNGNLEVSLFHGDYEVEISHPAKKNYTLTHHVQVVPNDESGKSS 122
Query: 583 QFIQLS 588
QF+QLS
Sbjct: 121 QFVQLS 104
>TC15537
Length = 589
Score = 51.6 bits (122), Expect = 4e-07
Identities = 37/112 (33%), Positives = 55/112 (49%), Gaps = 8/112 (7%)
Frame = +1
Query: 461 TELDVAS--QPNQALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLTDNNFKNLPAG 518
TELDV+S + +A E +LREA +HP ++GI++W W D++ G
Sbjct: 4 TELDVSSIYEHVRADDLEVMLREAMAHPAVEGIMLWGFWE-----LFMSRDHSHLVNAEG 168
Query: 519 DVVD------QLINEWGRAEKSGTTDQNGYFEASLFHGDYEIEINHPIKKKS 564
D+ + L EW + K D++G F FHG Y +E+ P KK S
Sbjct: 169 DINEAGKRFLDLKREW-LSHKHDHVDEHGQFSFRGFHGTYNVEVVTPTKKIS 321
>AW719969
Length = 263
Score = 26.9 bits (58), Expect = 9.3
Identities = 11/35 (31%), Positives = 19/35 (53%)
Frame = -1
Query: 185 WVDNVSLQPFTEKQWRSHQELSIEKDRKRKVVVRA 219
W++ L+ F E +W +L ++RKR + RA
Sbjct: 257 WLEEEGLREFEEVRWLRVLDLRTRRERKRVWITRA 153
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,676,956
Number of Sequences: 28460
Number of extensions: 146649
Number of successful extensions: 711
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 706
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 711
length of query: 588
length of database: 4,897,600
effective HSP length: 95
effective length of query: 493
effective length of database: 2,193,900
effective search space: 1081592700
effective search space used: 1081592700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146757.5


